Review Article
The Bacterial Heterotrimeric Amidotransferase GatCAB: functions, structures and mechanism-based inhibitors

Van Hau Pham* and Jacques Lapointe
Postdoctoral Researcher, Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, USA
*Address for Correspondence: Van Hau Pham, Postdoctoral Researcher, Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, USA, Tel: +1 317-658-0093; Email: [email protected]
Dates: Submitted: 06 April 2017; Approved: 28 April 2017; Published: 01 May 2017
How to cite this article: Pham VH, Lapointe J. The Bacterial Heterotrimeric Amidotransferase GatCAB: functions, structures and mechanism-based inhibitors. Arch Biotechnol Biomed. 2017; 1: 021-032. DOI: 10.29328/journal.abb.1001003
Copyright License: © 2017 Pham VH, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ABSTRACT
tRNA-dependent amidotransferases (AdT) are essential enzymes for protein biosynthesis in many bacteria and in all archaea. As AdT is essential for a number of pathogenic bacteria, and it is absent from mammalian cytoplasm, it is considered as a putative target for novel inhibitors that could be lead compounds to develop a new class of antibiotics. Besides GatFAB of Saccharomyces cerevisiae mitochondria and GatAB of Plasmodium falciparum apicoplast, all reported AdT can be divided into two groups: heterodimeric GatDE and heterotrimeric GatCAB. The latter is required to catalyze the conversion of Glu-tRNAGln and/or Asp-tRNAAsn into Gln-tRNAGln and/or Asn-tRNAAsn in many pathogenic bacteria. Recently determined high resolution crystal structures of several GatCAB could be used to design new inhibitors. In this review, we highlight the essential role of AdT for the faithful translation of glutamine and/or asparagine codons, we describe important features of the crystal structures of several GatCAB as well as tRNA/AdT/aaRS complexes for the formation of Gln-tRNAAsn and Asn-tRNAAsn, we finally summarize discoveries of AdT inhibitors based on their analogy to glutamine, adesosine tripoliphosphate and 3’-end of tRNA.
INTRODUCTION
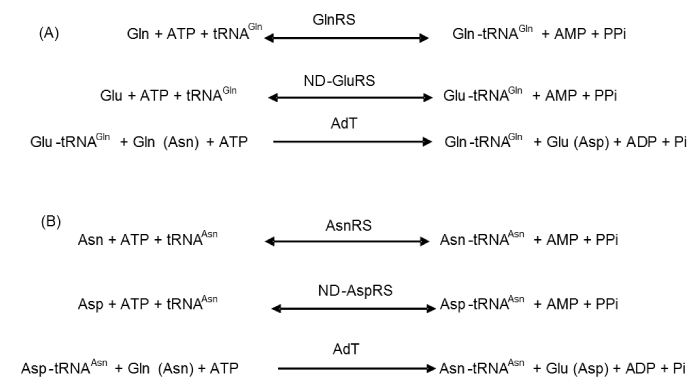
Accurate aminoacylation of tRNA is crucial for the faithful translation of genetic information into active and properly folded protein. In many organisms and in the cytoplasm of eukaryotic cells, each of about twenty aminoacyl-tRNA synthetizes recognizes and attaches its cognate tRNAs to its cognate amino acid (aa). The resulting products are the aminoacyl-tRNAs (aa-tRNA), which are then used by the ribosome for protein biosynthesis. However, most bacteria and eukaryotic organelles, and all archaea, lack the full set of 20 aminoacyl-tRNA synthetizes. For instance, Helicobacter pylori [1,2], Aquifex aeolicus [3] and Chlamydia trachomatis [1] do not have asparaginyl-tRNA synthetize (AsnRS) or glutaminyl-tRNA synthetase (GlnRS); Bacillus subtilis [4] and Staphylococcus aureus [5,6] are missing GlnRS, and Pseudomonas aeruginosa [1,7] lacks AsnRS. To survive, these micro-organisms must therefore use alternative pathways to generate asparaginyl-tRNAAsn (Asn-tRNAAsn) and glutaminyl-tRNAGln (Gln-tRNAGln). In these so-called indirect pathways, glutamate and aspartate are first attached to tRNAGln and tRNAAsn, respectively, in reactions catalyzed by a non-discriminating glutamyl-tRNA synthetize (ND-GluRS) and a non-discriminating aspartyl-tRNA synthetize (ND-AspRS). Then, the misacylated Glu-tRNAGln and Asp-tRNAAsn are converted into Gln-tRNAGln and Asn-tRNAAsn by a specific tRNA-dependent amidotransferase (AdT) [7,8] (Figure 1).
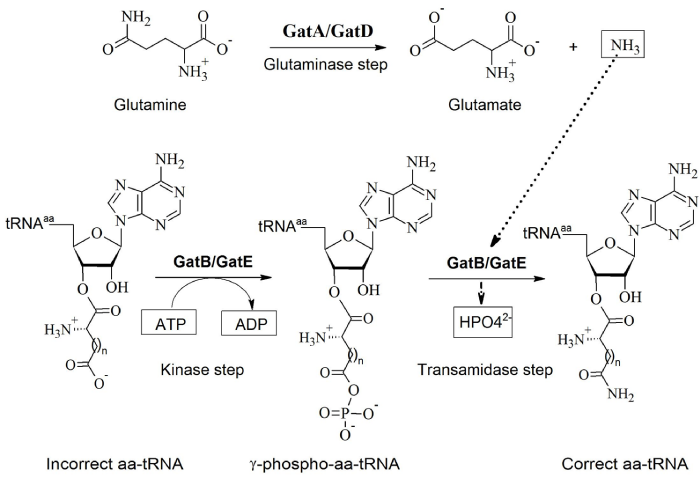
Two main types of AdT, heterotrimeric AdT (GatCAB) and heterodimeric AdT (GatDE) have been discovered so far. GatCAB, found in many bacteria, eukaryotic organelles and some archaea, can transamidate both Glu-tRNAGln and Asp-tRNAAsn. Unlike GatCAB, GatDE only exists in archaea and only transamidates Glu-tRNAGln, also called archaeal-specific AdT. The GatA and GatD subunits have a similar function and act as glutaminases, generating free ammonia from glutamine. GatB and GatE activate and transamidate the intermediate misacylated-tRNA to form the correctly charged tRNA species (Figure 2) [7]. GatC wraps around the GatA-GatB interface to correctly fold and to stabilize GatA and GatB [3,5,6,8]. Recently, two GatCAB-like AdT have been reported: GatFAB [9,10] and GatAB [11]. The former is found in mitochondria of Saccharomyces cerevisiae while the latter was discovered in Plasmodium falciparum apicoplasts. Although GatF does not share sequence similarity with GatC, the C-terminal of GatF plays a role similar to that of GatC, stabilizing the trimeric structure. The N-terminal part of GatF interacts with GatA and wraps around the glutaminase site. The GatF subunit is fungi-specific and twice longer than GatC [9].
Figure 2: Similar functions of the subunits of GatCAB and GatDE in the formation of Asn-tRNAAsn (when n=1) and Gln-tRNAGln (when n=2) (Adapted from [8]).
For the following reasons, targeting the GatCAB of pathogenic bacteria is a promising way to generate new antibiotics. Firstly, GatCAB is an essential enzyme for the growth of pathogenic bacteria such as H. pylori, S. aureus and P. aeruginosa; its inhibition may be lethal for these bacteria. Secondly, it is absent from mammalian cytoplasm, which precludes that bactericidal AdT inhibitors would necessarily have side-effects when introduced into clinical use. Thirdly, as this enzyme two catalytic centers binding four substrates, several types of substrate-based inhibitors can be considered, targeting the binding sites of ATP, glutamine, NH3 or aa-tRNA; moreover, from the present knowledge of the mechanisms of the reactions catalyzed by GatCAB, new reaction-intermediate-based inhibitors could also be designed.
In this review, we summarize the present knowledge about the structures and the functions of the heterotrimeric AdT. We also report the recent description of tRNA/AdT/aaRS complexes which promote the formation of Gln-tRNAGln and/or Asn-tRNAAsn. We finally highlight the findings of GatCAB inhibitors analogous to glutamine, ATP and the 3’-end of Glu-tRNAGln and Asp-tRNAAsn.
THE STRUCTURE AND FUNCTIONS OF GATCAB
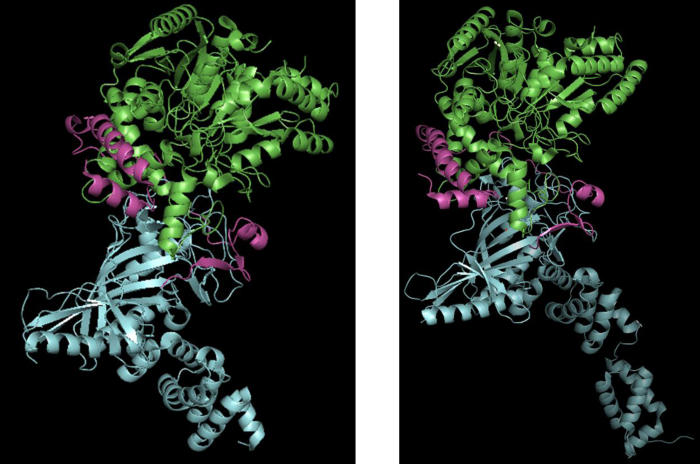
The crystal structures of the heterotrimeric AdT (or GatCAB) from S. aureus and A. aeolicus have been determined at high resolution [3,5,6]. The GatA and GatB subunits perform respectively functions as glutaminase and synthetase (which includes kinase and transamidase) [3], whereas the GatC subunit promotes the correct folding of GatA in the B. subtilis enzyme, the first GatCAB to be purified and mechanistically characterized [4]. Moreover, crystal structures of S. aureus and A. aeolicus AdTs show that GatC makes extensive interactions with GatA and GatB [3,6] (Figure 3).
Figure 3: Crystal structures of A. aeolicus GatCAB at 2.3 Å (PDB: 3H0L) (left) and S. aureus GatCAB at 1.9 Å (3IP4) (right). GatA, GatB and GatC are illustrated in green, blue and magenta, respectively. S. aureus GatB includes the tail domain which is not shown in A. aeolicus GatB.
The GatA subunit of A. aeolicus has 478 a residues, is composed of an 11-stranded β-sheet surrounded by 12 α-helices, and includes a Ser-cisSer-Lys catalytic scissor active site formed by Ser171, Ser147, and Lys72 [3,12]. S. aureus GatA has a similar 3D-structure with 11 β-strands, 15 α-helices [6] and also a Ser-CisSer-Lys catalytic triad including residues Ser178, Ser154 and Lys79. The NH3 produced in both species by the GatA glutaminase active site, is thought to reach the GatB active site via a channel through GatA and GatB [3,6].
The GatB subunit of both microorganisms (478 aa) is made from an N-terminal “cradle” domain and a helical domain followed by a highly flexible C-terminal domain [6]; in a crystal obtained under slightly modified conditions, and diffracting at higher resolution, the structure of this “tail domain” in S. aureus GatCAB was determined [5]. Its structure is similar to that of the YqeY-like domain, which when appended to D. radiodurans GlnRS, enables this enzyme to productively bind to tRNAGln [13]. The cradle domain is capable of coordinating two divalent metal ions which form two distinct active sites on GatB, kinase and transamidase, performing together the synthetase function (see above). The kinase active site of A. aeolicus GatCAB was identified from the position of ATP bound to one of the Mn2+ of the cradle domain of GatB, in a crystal of the GatCAB/ATP complex quick-frozen in liquid N2; its adenine base sits in the hydrophobic pocket formed by Val8, Phe208 and Pro158 [3], and makes hydrogen bonds with its N1 and N6 atoms to the conserved Ser199 side chain. ADP, a product of the hydrolyzed ATP, was also found at the same site in another similar crystal not quick-frozen in liquid N2, confirming the precise binding site of ATP. The ADP α- and β-phosphates make interactions with bound water molecules, whereas the ATP γ-phosphate coordinates with a metal ion (thought to be Mg2+ in vivo) in a transient binding site. This transient metal ion is bound to the conserved residues Glu12 and Glu213 of A. aeolicus GatB and to one water molecule. Its role is to assist phosphoryl transfer by polarizing the γ-phosphate group of ATP, provoking the nucleophile attack to form γ-phosphoryl-Glu-tRNAGln or β-phosphoryl-Asp-tRNAAsn [3,5,6]. In the transamidase active site of A. aeolicus GatB, another metal ion binding site, called permanent due to its retaining a metal ion in all the crystal structures of A. aeolicus GatCAB examined (5), is fixed on GatB by coordination with His14, Glu127 and Glu153 [3]. Crystallizing GatCAB in the presence of Asp, a mimic of the substrate Asp-tRNAAsn, showed that the Asp electron density was close to the permanent Mn2+. Interestingly, one of the carboxyl groups of Asp was found to coordinate the metal ion, and it faces the ATP γ-phosphate. By interacting with the variable loop and D-loop of tRNA, the tail domain is partially responsible for distinguishing tRNAGln from tRNAGlu [5].
The GatC subunit (94 residues) is four times smaller than GatA or GatB. Its role is to stabilize GatA as well as the trimeric structure [3,5,6].
The indirect formation of Gln-tRNAGln and/or Asn-tRNAAsn requires the participation of three macromolecules: a ND-aaRS (or a noncanonical aaRS such as GluRS2), an AdT and a tRNA. The term “transamidosome” has been literally used so far to describe either a ternary complex or a binary complex. The term “transamidosome” was first defined as a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis [14] to indicate a ternary complex composed of tRNA, aaRS and AdT. However, this term was also used for ternary complexes without tRNA, such as the H. pylori Asn-transamidosome containing Hp0100, AdT and GatCAB [15], or for the binary complexes formed by AdT and aaRS, such as the Methanobacter thermautotrophicus Gln-transamidosme [16]; in the latter cases, tRNA does not play the role of a scaffold to assist the assembly of the ternary complexes, but only serves as a substrate. The transamidosomes of five micro-organisms involved in Gln-tRNAGln and Asn-tRNAAsn formation have been structurally described and/or biochemically characterized [14,15,17-19]. The structures and the assembly mechanisms of these characterized transamidosomes are different from one another, but they all catalyze the formation of Gln-tRNAGln and Asn-tRNAAsn by either binary or ternary complexes (Table 1).
| Table 1: Structures and characteristics of the known Asn- and Gln-transamidosomes. | |||||
| Organisms | Methods used to characterize the transamidosome | Characteristics of transamidosomes | References | ||
| Type of transamidosome and its components | Stoichiometry (aaRS/tRNA/AdT) | Stability | |||
| Thermus thermophilus | X-ray crystallography SAXS Gel filtration Dynamic light scattering |
Archeal-type, Asn-transamidosome tRNA, ND-AspRS, GatCAB | 2/4/4 in crystal 1/2/2 in solution |
Stable over the entire catalytic cycle | [14,17] |
| Thermotoga maritima | X-ray crystallography | Bacterial-type, Gln-transamidosome tRNA, GluRS, GatCAB | 1/2/1 in crystal | Dynamic | [23] |
| Methanothermobacter thermautotrophicus | Gel filtration | Archeal-type, Gln-transmidosome ND-GluRS, GatDE | Not reported | Stable, even in the absence of tRNA | [16] |
| Helicobacter pylori | Dynamic light scattering | Bacterial-type, Asn-transamidosome Hp0100, ND-AspRS, GatCAB | Not determined | Stable only in the presence of Hp0100 | [15] |
| Helicobacter pylori | Gel filtration Dynamic light scattering |
Bacterial-type, Gln-transamidosome tRNA, GluRS2, GatCAB | 1/1/1 in solution | Dynamic in the absence of tRNA; and stable in its presence | [19] |
| Helicobacter pylori | Gel filtration Dynamic light scattering |
Bacterial-type, Asn-transamidosome tRNA, GluRS2, GatCAB | Not reported | Dynamic in the absence of tRNA; and stable in its presence | [24] |
| Pseudomonas aeruginosa | X-ray crystallography | Bacterial-type, Asn-transamidosme tRNA, ND-AspRS, GatCAB | 2/2/2 in crystal | Dynamic in the absence of tRNA; and stable in its presence | [20] |
In most of the transamidosomes, involving GatCAB, ND-aaRS first binds tRNA. Then, the GatB tail-domain binds this binary complex via the D-loop of tRNA. The product of the aminoacylation reaction, Asp-tRNA or Glu-tRNA, triggers conformational changes on the ND-aaRS which bring about the release of the tRNA acceptor stem. Immediately, this stem flips up toward GatCAB [20,21]. The transamidation then takes place on the ternary complex, using ammonia generated from GatA to form the correct aa-RNA (Gln-tRNAGln or Asn-tRNAAsn).
The crystal structure of the T. thermophilus transamidosome was first determined at 3 Å resolution, showing a 520 kDa particle formed by two GatCAB, two dimeric ND-AspRS and four tRNAAsn [17]. Only two tRNA molecules were found in the GatB active site; the two other tRNAs acted as a scaffold to stabilize the complex. In solution, this ternary structure was comprised of one dimeric ND-AspRS, two GatCABs and two tRNAs (380 kDa), consistent with the complex previously characterized by gel filtration [14]. The structures of ND-AspRS in the complex were similar to those described for other bacterial and eukaryotic counterparts [22,23]. GatCAB also resembles to those individually characterized in A. aeolicus and S. aureus [3,5,6]. It was suggested that this transamidosome prevents the release of mischarged aa-tRNA, thus preventing its interaction with EF-Tu and the ribosome, and that it protects GatCAB from heat denaturation at 85oC, the optimal growth temperature of this bacterium [17].
The formation of Gln-tRNAGln in M. thermoautotrophicus also relied on a transamidosome [16]. Unlike what was found for the T. thermophilus transamidosome, the M. thermoautotrophicus transamidosome contains only two components: ND-GluRS and GatDE. The GatDE/ND-GluRS binary complex was formed first, and then recruited tRNA to generate Gln-tRNAGln. Detected by filtration on a Sephacryl S300 gel, this archaeal GatDE/ND-GluRS complex migrated as a particle of 380 kDa, suggesting the association of a GatDE homodimer with two ND-GluRS monomers. Nevertheless, 3D structures have not been reported for this archaeal transamidosome of M. thermoautotrophicus.
The structures of the GluRS/tRNAGln/GatCAB ternary and GluRS/tRNAGln binary complexes of Thermotoga maritima were determined by X-ray crystallography and gel mobility shift assay [23]. This bacterial Gln-transamidosome was described as the assembly of tRNAGln, GluRS and GatCAB. In this transamidosome, one hinge in GluRS and two hinges in GatB, ensure the flexibility for the movement of the a acceptor arm of tRNA from the GluRS active site to the GatB active site. In the crystal structure of the T. maritima transamisdosome, the acceptor arm of tRNA intrudes into the catalytic site of GluRS, which was not the case for the T. thermophilus [17] and the P. aeruginosa Asn-transamidosomes [20] in which the a acceptor arm of tRNAGln pointed toward the cradle domain of the GatB catalytic site. These results suggest that the acceptor arm of tRNA in transamidosomes must move from the ND-aaRS active site to the GatB transamidation active site.
For H. pylori, a unique GatCAB catalyzes the conversion of both Asp-tRNAAsn and Glu-tRNAGln into Asn-tRNAAsn and Gln-tRNAGln, respectively, but there are two distinct transamidosomes. In the H. pylori Asn-transamidosome, a specific protein named as Hp0100 is required for the transamidosome formation [15]. This transamidosome was described as an assembly of a ND-AspRS, GatCAB and Hp0100, and did not require tRNAAsn. Indeed, protein Hp0100 which is an auxiliary factor, assists the association of ND-AspRS and GatCAB in the absence of tRNA. Unlike the H. pylori Asn-transamidosome, the H. pylori Gln-transamidosome revealed a particle which contained one non-canonical GluRS2, one tRNA and one GatCAB. tRNA played a role as a scaffold molecule for the formation of the ternary complex, with an optimum at equal molarity to those of the two other macromolecules (GluRS2 and GatCAB). Intriguingly, when the tRNA concentration was higher than that optimal for complex formation, the transamidosome separated into GluRS2/tRNA and GatCAB/tRNA binary complexes [19], indicating that the transamidosome is dynamic and less stable than the T. thermophilus Asn-transmidosome and the M. thermoautotrophicus Gln-transamidosome. Glu-tRNA and Gln-tRNA were reported to be protected from the hydrolysis within this transamidosome, suggesting that the complex ensures the correct translation of Gln codons in protein biosynthesis in H. pylori.
Most recently, the structure of the P. aeruginosa Asn-transamidosome was reported at 3.7 Å resolution. Its structure is a symmetric complex including two ND-AspRS, two GatCAB and two tRNAAsn (approximately 400 kDa) [18,20]. Differing from the T. thermophilus archaeal-type transamidosome, the P. aeruginosa complex possesses a GAD domain in the ND-AspRS, which is specific for bacterial-type transamidosomes [20], and both tRNAs are catalytically active. Similar to what was reported for the T. thermophilus Asn-transamidosome, the acceptor arm of tRNA in the P. aeruginosa Asn-transamidosome binds to the GatB transamidation active site. The crystal structure showed few interactions between GatB and ND-AspRS, which do not form a stable binary complex, underlining the requirement of tRNA for the assembly of the P. aeruginosa Asn-transamidosome [20].
Overall 3D-structures as well as biochemical characterization of the known Asn- and Gln-transamidosomes deepen substantial understandings of their functions and reaction mechanisms. These described transamidosomes have different stoichiometries, modes of assembly and positions of the tRNA acceptor arm. Intriguingly, the H. pylori Asn-transamidosome required Hp0100, an auxiliary protein factor specific to ɛ-proteobacteria. These differences can be exploited to discover specific inhibitors according to proper characteristics in each microorganism as suggested for clade-specific antibiotics targeting the assembly of the H. pylori Asn-transamidosome [15]. Lastly, the future antibiotics could act on the movement of the tRNA acceptor arm from ND-GluRS2 toward GatCAB, and/or on the active sites of GatCAB for the pathogenic bacterium H. pylori.
MECHANISM-BASED INHIBITORS OF GATCAB
GatCAB inhibitors analogous to glutamine
In 2001 and 2002, a group led by Robert A. Copeland at Dupont Pharmaceuticals Company, found the first inhibitors of AdT, targeting the glutaminase active site of Streptococcus Pyogenes GatA, and also demonstrated the involvement of residue Ser176 in this site [25-27]. Decicco et al. (2001) synthesized a series of analogues of glutamyl-γ-boronate, as mechanism-based inhibitors of bacterial AdT designed to engage a putative catalytic serine nucleophile required for the glutaminase activity of the enzyme. These compounds, which inhibit S. pyogenes AdT [25], were synthesized by the replacement of the γ-carboxamide of glutamine with boronic acid. The authors assumed that the conserved Ser176 of S. pyogenes GatA involved in the catalytic reaction as a nucleophile, was neutralized by the boronic acid component (electrophile) and eventually formed a serine-boronate acetal. The three strongest inhibitors of AdT, among these 21 analogues of glutamyl-γ-boronate, displayed IC50 values from 1.3 to 1.6 µM against the glutaminase activity, and from 50 to 100 nM against the transferase activity. In vivo, these compounds inhibit the growth of S. pyogenes, S. pneumonia, and Enterobacter faecalis but not that of S. aureus and H. pylori, despite the fact that these two organisms have an AdT. By replacing Ser176 by Ala in S. pyogenes GatA, Harpel et al. [26] demonstrated that this serine is essential for the cleavage of Gln into Glu and NH3. This residue uses its hydroxyl group as a nucleophile to facilitate the catalytic reaction. γ-Glu boronic acid was considered as a nucleophile trap and shown to be able to trap Ser176. Harpel et al. [26] also demonstrated that γ-Glu boronic acid is a competitive inhibitor of AdT with respect to glutamine, with a Ki value of 73 nM [26]. The involvement of Ser176 in glutamine hydrolysis remained putative until the structures of S. aureus and of A. aeolicus AdT revealed that their GatA residues (Ser178 and Ser171, respectively) corresponding to Ser176 in S. pyogenes GatA, are part of its glutaminase catalytic site.
GatCAB inhibitors analogous to ATP
Horiuchi et al. [27] reported evidence of tight kinetic coupling between the glutaminase, transamidase and ATP hydrolysis activities of S. pyogenes AdT. In the absence of the amido acceptor, Glu-tRNAGln, this AdT has basal glutaminase activity that is unaffected by ATP. However, Glu-tRNAGln activates the glutaminase activity of the enzyme about 10-fold; addition of ATP elicits a further 7-fold increase. To determine if ATP binding is sufficient to induce full activation, they tested a variety of ATP analogues for their ability to stimulate tRNA-dependent glutaminase activity. Among these analogues, only ATP-γS stimulates glutaminase activity, and does it to the same level as ATP. This analogue is also a weak inhibitor (IC50=0.19 mM) of the transamidase activity, and a weak activator (3-fold) of the glutaminase activity.
GatCAB inhibitors analogous to the 3’-end of Glu-tRNA or Asp-tRNA
The above-mentioned AdT inhibitors, analogous to glutamine or ATP, are likely to inhibit numerous enzymes which use glutamine or ATP as substrates. On the other hand, small molecules analogous to the 3’-end of one of the aa-tRNA substrates of AdT, Glu-tRNA or Asp-tRNA, are more likely to target AdT specifically.
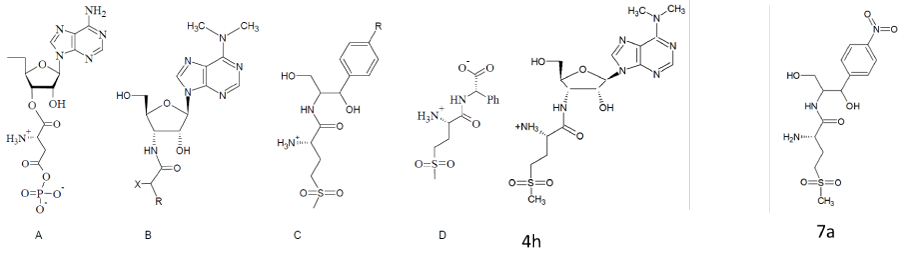
Glutamycin, a stable analogue of the 3’-end of Glu-tRNA, initially shown to be a weak inhibitor of Glu-tRNA reductase [28], and its close analogue aspartycin were the first known inhibitors designed against the synthetase site of AdT [29]. They are competitive inhibitors of H. pylori GatCAB with respect to it aa-tRNA substrate, and exhibit Ki values of 134 µM for aspartycin and 105 µM for glutamycin, respectively [29]. Analogues of tetrahedral intermediates formed transiently during the transamidation reaction, and analogues of the aa-tRNA products were identified as stronger inhibitors than glutamycin and aspartycin [29]; among them, the sulfur-containing puromycin derivatives are the strongest AdT inhibitors, the best being the sulfone “4h” compound with a Ki value of 4 µM [30]. Puromycin is an aminonucleoside antibiotic produced by Streptomyces alboniger, which mimics the charged 3’-terminus of aminoacylated tRNA, and accepts peptide chains from peptidyl-tRNA on the ribosome. It is a very weak inhibitor of H. pylori AdT GatCAB (Ki=4 mM), probably because the methoxyphenyl moiety of puromycin is related to tyrosine and not to the aspartic and glutamic side chains which are transformed by AdT. Replacement of this moiety by carboxylic acid derivatives gave analogues of the 3’-ends of Asp-tRNA and Glu-tRNA, which are much better inhibitors of AdT (Ki values about 100 µM) and are competitive with respect to the H. pylori Asp-tRNAAsn substrate [30].
Other strong inhibitors, also competitive with respect to Asp-tRNA, were obtained by effectively replacing the puromycin aminonucleoside module of the above described AdT inhibitors with a chloramphenicol-like module [31]. The antibiotic chloramphenicol inhibits protein synthesis by binding to the peptidyl transferase region of the ribosome, and overlaps the binding site of puromycin [32,33]. The similarity of these two compounds led [31] to explore the potential of L-methionine-sulfone derivatives of chloramphenicol as AdT inhibitors, a strategy based on the premise that the sulfone moiety mimics the transition state in the transamidation reaction (last step in Figure 2). Whereas chloramphenicol is a very weak inhibitor of H. pylori GatCAB (Ki=1.9 mM), replacement of its dichloroacetyl moiety by several L-methionyl-sulfone analogs considerably enhanced the inhibitory activity against this enzyme ; among them, compound 7a is the best inhibitor with a Ki value of 27 µM, and it is competitive with respect to H. pylori Asp-tRNAAsn (Figure 4).
Figure 4: Strategy for the design of GatCAB inhibitors. A: putative intermediate of the transamidation reaction, B: puromycin analogues, C: chloramphenicol analogues, D: truncated forms of the compound 4h, 4h: the structures of the best puromycin analogues and 7a: the structure of the best chloramphenicol analogues.
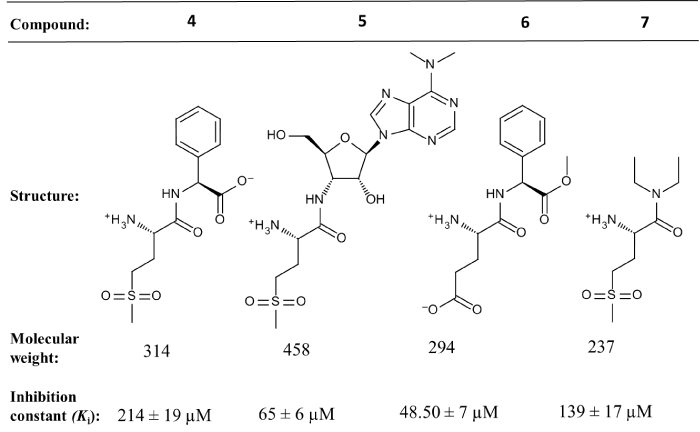
Recently, we have reported the inhibition properties of the truncated derivatives of the best inhibitor “4h”, named as compound 5 (Figure 5). These compounds, lacking the bulky module, are analogues of the transition state of the transamidation reaction, are competitive inhibitors of AdT with respect to its aa-tRNA substrate, with Ki from 48.50 to 214 μM [34] and figure 5. Their properties reveal that the 3’-terminal adenine of tRNA plays a major role in binding the 3’-end of Glu-tRNAGln into the AdT synthetase active site.
Figure 5: Characteristics of the transition state analogues of the putative transamidation reaction catalyzed by AdT.
The synthesis of non-hydrolyzable analogs of the 3’-end of for Asp-tRNA or Glu-tRNA was reported by Klinchan et al. but no information on their inhibitory action on AdT was included [35]. Recently, Pham et al. have published four cyclic peptides inhibiting H. pylori GatCAB [36]. These peptides which are rich in tryptophan and proline were discovered by phage display technique. Two peptides P10 (CMPVWKPDC) and P9 (CSAHNWPNC) are competitive inhibitors with Ki values of 126 and 392 µM, respectively, with respect to Glu-tRNAGln. The docking models of these two peptides into H. pylori subunit B showed that they bound to the trasmidation active site of GatB via π-π interactions with Tyr81, as does the 3’-terminal A76 of tRNA.
DISCUSSION & CONCLUSION
The glutaminase activity of S. pyogenes Glu-tRNAGln amidotransferase (Glu-AdT) is efficiently inhibited by some analogues of glutamyl-γ-boronate [26]. The bests of these mechanisms-based inhibitors, with IC50 values of about 1.5 µM against the glutaminase activity of S. pyogenes Glu-AdT, also inhibit efficiently the growth of this bacteria as well as that of other bacteria that have an AdT (Streptococcus pneumoniae and Enterobacter faecalis) (MIC of 2 to 8 μ/ml), but do not inhibit significantly the growth of other bacteria that have an AdT (S. aureus and H. pylori), and do not inhibit the growth of Escherichia coli, which does not have an AdT. These results validate mechanism-based inhibitor design for Glu-AdT as an approach to antimicrobial development, but such compounds are likely to inhibit numerous enzymes which use glutamine as substrates.
The influence of various ATP analogues on S. pyogenes Glu-AdT was tested by Horiuchi et al. [28] in their study of the tight kinetic coupling between the glutaminase, transamidase and ATP hydrolysis activities of this enzyme; none of these compounds have a significant inhibitory activity.
Several small compounds, stable analogues of the 3’-end of Glu-tRNA or of Asp-tRNA, were designed from the structures of puromycin and of chloramphenicol [31,32], two antibiotics which interfere with peptide bond formation on prokaryotic ribosomes [33,34]. Their stability is due to an amide bond between the module analogous to the aminoacyl group of aa-tRNA and the module analogous to the 3’-terminal A76 of tRNA. By contrast, the “high energy” ester bond at the corresponding position in aa-tRNA is unstable, and its half-life depends on the nature of the amino acid and on the ionic strength (reviewed by Söll and Schimmel [37]). In vivo, this aa-tRNA ester bond is stabilized by its interaction with the elongation factor (EF-Tu in bacteria) [38] which is present at high concentrations in living cells [39]. The instability of this ester bond in the AdT substrates Glu-tRNA and Asp-tRNA is a major obstacle for the identification by X-ray crystallography of the binding site for these tRNA-linked amino acids on AdT, as the time required to grow crystals of AdT/aa-tRNA complexes is probably too long to keep this ester bond; for instance, the half-life of H. pylori Glu-tRNAGln is about 4 hours at pH 7.2 in the presence of H. pylori GatCAB, and about 1 hour in its absence [19]. Recently-reported Gln- and Asn-transamidosomes depicted that the acceptor arm CCA of tRNA flips from the active site of aaRS toward the transamidation active site of the cradle domain of GatB, suggesting that tRNA is dynamic in order to simulatiously be accommodated into these two active sites.
The above-mentioned small puromycin analogues, which are competitive inhibitors of AdT with respect to its aa-tRNA substrate, are probes for further studies of AdT mechanism, and are ligands for X-ray crystallographic studies of the AdT synthase active site. Moreover, as this indirect pathway for the formation of Gln-tRNAGln and Asn-tRNAAsn, involving ND-GluRS and/or ND-AspRS and AdT, is present in many pathogenic bacteria, these small mechanism-based inhibitors open potential avenues to develop antibiotics with a novel mode of action.
REFERENCES
- Sheppard K, Akochy PM, Salazar JC, Söll D. The Helicobacter pylori amidotransferase GatCAB is equally efficient in glutamine-dependent transamidation of Asp-tRNAAsn and Glu-tRNAGln. J Biol Chem. 2007; 282: 11866-11873. Ref.: https://goo.gl/dfgopk
- Chang KM, Hendrickson TL. Recognition of tRNAGln by Helicobacter pylori GluRS2--a tRNAGln-specific glutamyl-tRNA synthetase. Nucleic Acids Res. 2009; 37: 6942-6949. Ref.: https://goo.gl/vFSjrB
- Wu J, Bu W, Sheppard K, Kitabatake M, Kwon ST, et al. Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme. J Mol Biol. 2009; 391: 703-716. Ref.: https://goo.gl/0IEr7c
- Curnow AW, Hong Kw, Yuan R, Kim Si, Martins O, et al. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci U S A, 1997; 94: 11819-11826. Ref.: https://goo.gl/VtHKqn
- Nakamura A, Sheppard K, Yamane J, Yao M, Söll D, et al. Two distinct regions in Staphylococcus aureus GatCAB guarantee accurate tRNA recognition. Nucleic Acids Res. 2010; 38: 672-682. Ref.: https://goo.gl/Oz1r7J
- Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia Channel Couples Glutaminase with Transamidase Reactions in GatCAB. Science. 2006; 312: 1954-1958. Ref.: https://goo.gl/VTTBFo
- Sheppard K, Yuan J, Hohn MJ, Jester B, Devine KM, et al. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008; 36: 1813-1825. Ref.: https://goo.gl/i0x0fN
- Huot JL, Jacques Lapointe, Robert Chênevert, Marc Bailly, Daniel Kern. 5.14-Glutaminyl-tRNA and Asparaginyl-tRNA Biosynthetic Pathways. Elsevier. 2010; 383-431. Ref.: https://goo.gl/ZwhxGb
- Araiso Y, Huot JL, Sekiguchi T, Frechin M, Fischer F, et al. Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases. Nucleic Acids Res. 2014; 42: 6052-6063. Ref.: https://goo.gl/pZmBNq
- Frechin M, Senger B, Brayé M, Kern D, Martin RP, et al. Yeast mitochondrial Gln-tRNA(Gln) is generated by a GatFAB-mediated transamidation pathway involving Arc1p-controlled subcellular sorting of cytosolic GluRS. Genes Dev. 2009; 23: 1119-1130. Ref.: https://goo.gl/QqkouY
- Mailu BM, Arthur J, Nelson TM, Ramasamy G, Fritz-Wolf K, et al. Plasmodium Apicoplast Gln-tRNAGln Biosynthesis Utilizes a Unique GatAB Amidotransferase Essential for Erythrocytic Stage Parasites. J Biol Chem, 2015; 290: 29629-29641. Ref.: https://goo.gl/gaHBGQ
- Shin S, Yun YS, Koo HM, Kim YS, Choi KY, et al. Characterization of a Novel Ser-cisSer-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad. J Biol Chem. 2003; 278: 24937-24943. Ref.: https://goo.gl/lkXkIV
- Deniziak M, Sauter C, Becker HD, Paulus CA, Giegé R, et al. Deinococcus glutaminyl-tRNA synthetase is a chimer between proteins from an ancient and the modern pathways of aminoacyl-tRNA formation. Nucleic Acids Res. 2007; 35: 1421-1431. Ref.: https://goo.gl/QHkb2F
- Bailly M, Blaise M, Lorber B, Becker HD, Kern D. The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol Cell. 2007; 28: 228-239. Ref.: https://goo.gl/yYxtDv
- Silva GN, Fatma S, Floyd AM, Fischer F, Chuawong P, et al. A tRNA-independent mechanism for transamidosome assembly promotes aminoacyl-tRNA transamidation. J Biol Chem. 2013; 288: 3816-3822. Ref.: https://goo.gl/MTFWiy
- Rampias T, Sheppard K, Soll D. The archaeal transamidosome for RNA-dependent glutamine biosynthesis. Nucleic Acids Res. 2010; 38: 5774-5783. Ref.: https://goo.gl/ed5tXy
- Blaise M, Bailly M, Frechin M, Behrens MA, Fischer F, et al. Crystal structure of a transfer-ribonucleoprotein particle that promotes asparagine formation. EMBO J. 2010; 29: 3118-3129. Ref.: https://goo.gl/f5bUIx
- Suzuki T, Yamashita K, Tanaka Y, Tanaka I, Yao M. Crystallization and preliminary X-ray crystallographic analysis of a bacterial Asn-transamidosome. Acta Crystallogr F Struct Biol Commun. 2014; 70: 790-793. Ref.: https://goo.gl/KzGbS9
- Huot JL, Fischer F, Corbeil J, Madore E, Lorber B, et al. Gln-tRNAGln synthesis in a dynamic transamidosome from Helicobacter pylori, where GluRS2 hydrolyzes excess Glu-tRNAGln. Nucleic Acids Res. 2011; 39: 9306-9315. Ref.: https://goo.gl/w0Z72h
- Suzuki T, Nakamura A, Kato K, Söll D, Tanaka I, et al. Structure of the Pseudomonas aeruginosa transamidosome reveals unique aspects of bacterial tRNA-dependent asparagine biosynthesis. Proc Natl Acad Sci U S A. 2015; 112: 382-387. Ref.: https://goo.gl/rtlp1N
- Ito T, Yokoyama S. Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions. Nature. 2010; 467: 612-616. Ref.: https://goo.gl/K9F8jz
- Delarue M, Poterszman A, Nikonov S, Garber M, Moras D, et al. Crystal structure of a prokaryotic aspartyl tRNA-synthetase. EMBO J. 1994; 13: 3219-3229. Ref.: https://goo.gl/nMrB2j
- Ruff M, Krishnaswamy S, Boeglin M, Poterszman A, Mitschler A, et al. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991; 252: 1682-1689. Ref.: https://goo.gl/cbVSUq
- Fischer F, Huot JL, Lorber B, Diss G, Hendrickson TL, et al. The asparagine-transamidosome from Helicobacter pylori: a dual-kinetic mode in non-discriminating aspartyl-tRNA synthetase safeguards the genetic code. Nucleic Acids Res. 2012; 40: 4965-4976. Ref.: https://goo.gl/DqM04x
- Decicco CP, Nelson DJ, Luo Y, Shen L, Horiuchi KY, et al. Glutamyl-γ-boronate Inhibitors of Bacterial Glu-tRNAGln Amidotransferase. Bioorg Med Chem Lett. 2001; 11: 2561-2564. Ref.: https://goo.gl/LsO5SH
- Harpel MR, Horiuchi KY, Luo Y, Shen L, Jiang W, et al. Mutagenesis and mechanism-based inhibition of Streptococcus pyogenes Glu-tRNAGln amidotransferase implicate a serine-based glutaminase site. Biochemistry. 2002; 41: 6398-6407. Ref.: https://goo.gl/d60tIG
- Horiuchi KY, Harpel MR, Shen L, Luo Y, Rogers KC, et al. Mechanistic studies of reaction coupling in Glu-tRNAGln amidotransferase. Biochemistry. 2001; 40: 6450-6457. Ref.: https://goo.gl/o5rygn
- Moser J, Lorenz S, Hubschwerlen C, Rompf A, Jahn D. Methanopyrus kandleri glutamyl-tRNA reductase. J Biol Chem. 1999; 274: 30679-30685. Ref.: https://goo.gl/3Qr3zM
- Huot JL, Balg C, Jahn D, Moser J, Emond A, et al. Mechanism of a GatCAB amidotransferase: aspartyl-tRNA synthetase increases its affinity for Asp-tRNA(Asn) and novel aminoacyl-tRNA analogues are competitive inhibitors. Biochemistry. 2007; 46: 13190-13198. Ref.: https://goo.gl/4TsUWh
- Balg C, Huot JL, Lapointe J, Chenevert R. Inhibition of Helicobacter pylori aminoacyl-tRNA amidotransferase by puromycin analogues. J Am Chem Soc. 2008; 130: 3264-3265. Ref.: https://goo.gl/zBSNkv
- Balg C, De Mieri M, Huot JL, Blais SP, Lapointe J, et al. Inhibition of Helicobacter pylori aminoacyl-tRNA amidotransferase by chloramphenicol analogs. Bioorg Med Chem. 2010; 18: 7868-7872. Ref.: https://goo.gl/Yy2Kd4
- Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001; 413: 814-821. Ref.: https://goo.gl/56Oe4A
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000; 289: 920-930. Ref.: https://goo.gl/MSrl7m
- Pham VH, Maaroufi H, Balg C, Blais SP, Messier N, et al. Inhibition of Helicobacter pylori Glu-tRNAGln amidotransferase by novel analogues of the putative transamidation intermediate. FEBS Lett. 2016; 590: 3335-3345. Ref.: https://goo.gl/N6HIkv
- Klinchan C, Yu-Ling H, Chiang LL, Pluempanupat W, Chuawong P. Synthesis of non-hydrolyzable substrate analogs for Asp-tRNAAsn/Glu-tRNAGln amidotransferase. Tetrahedron Letters. 2014; 55: 6204-6207. Ref.: https://goo.gl/j5Gn5W
- Pham VH, Maaroufi H, Levesque RC, Lapointe J. Cyclic peptides identified by phage display are competitive inhibitors of the tRNA-dependent amidotransferase of Helicobacter pylori. Peptides. 2016; 79: 8-15. Ref.: https://goo.gl/1Oej07
- Söll D, Schimmel PR. 15.Aminoacyl-tRNA Synthetases. The Enzymes. 1974; 489-538. Ref.: https://goo.gl/mY4Bj9
- Nissen P, Thirup S, Kjeldgaard M, Nyborg J. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure. 1999; 7: 143-156. Ref.: https://goo.gl/0NwJ0J
- Stepanov VG, Nyborg J. Thermal stability of aminoacyl-tRNAs in aqueous solutions. Extremophiles. 2002; 6: 485-490. Ref.: https://goo.gl/JquM6u