More Information
Submitted: 14 October 2020 | Approved: 26 October 2020 | Published: 28 October 2020
How to cite this article: Alzahrani NH, Mohamed EA. Evaluation of the antibacterial and anticancer activities of marine Bacillus subtilis ESRAA3010 against different multidrug resistant Enterococci (MDRE) and cancer cell lines Arch Biotechnol Biomed. 2020; 4: 018-027.
DOI: 10.29328/journal.abb.1001018
ORCiD: orcid.org/0000-0001-5168-5685
Copyright License: © 2020 Alzahrani NH, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Multidrug resistant Enterococci; Marine Bacillus; Antibacterial; Anti-cancer; Molecular identification
Evaluation of the antibacterial and anticancer activities of marine Bacillus subtilis ESRAA3010 against different multidrug resistant Enterococci (MDRE) and cancer cell lines
Nourah Hassan Alzahrani and Esraa Ahmed Mohamed2*
1Department of Biology, College of Science, University of Jeddah, Jeddah, Saudi Arabia
2Faculty of Medicine, October 6 University, 6th of October City, Giza, Egypt
*Address for Correspondence: Esraa Ahmed Mohamed, Faculty of Medicine, October 6 University, 6th of October City, Giza, Egypt, Email: [email protected]
Fifty nine isolates belonging to six species of Enterococci namely, Enterococcus faecalis, Enterococcus faecium, Enterococcus raffinosus, Enterococcus durans, Enterococcus mundtiiand Enterococcus avium (n = 35, 15, 4, 3, 1 and 1 isolates, respectively) were obtained from different clinical specimens including urine, pus, blood, wound, sputum and synovial fluid. The highest numbers of Enterococci were recorded from the pus (20 isolates, 33.90%) followed by urine (12 isolates, 20.34%) while the lowest frequency was observed with synovial fluid samples (2 isolates, 3.39%). These isolates showed different multidrug resistant patterns with the lowest resistant for linezolid (n = 5, 8.48%), followed by teicoplanin (n = 14, 23.73%) and vancomycin (n = 20, 33.90%) while they exhibited the highest resistant against penicillin (n = 53, 89.83%), oxacillin (n = 50, 84.75%), erythromycin (n = 49, 83.05%) and streptomycin (n = 47, 79.66 %). On the other hand, a free living marine bacterium under isolation code ESRAA3010 was isolated from seawater samples obtained from the fishing area Masturah, Red Sea, Jeddah, Saudi Arabia. The phenotypic, chemotaxonomic, 16S rRNA gene analyses and phylogenetic data proved that isolate ESRAA3010 is very close to Bacillus subtilis and then it was designated as Bacillus subtilis ESRAA3010. It gave the highest antagonistic activity against all clinical Enterococcus faecalis, Enterococcus faecium, Enterococcus raffinosus, Enterococcus durans, Enterococcus mundtiiand Enterococcus avium isolates under study with minimum inhibitory concentration (MIC) ranged from 4 to 56 µg/mL, 4 to 12 µg/mL, 4 to 8 µg/mL, 4 to 8 µg/mL, 8 µg/mL and 4 µg/mL, respectively as well as minimum bactericidal concentration (MBC) (8 to 64 µg/mL, 4 to 16 µg/mL, 4 to 12 µg/mL, 4 to 16 µg/mL, 12 µg/mL and 8 µg/mL, respectively). Moreover it showed anti-proliferative activity against colon (HCT-116), liver (HepG-2), breast (MCF-7) and lung (A-549) carcinomas with IC50 equal to 39, 50, 75 and 19 µg/mL, respectively which indicates its prospective usage in the upcoming decades.
Enterococcus species are Gram-positive cocci usually present in the human and sometimes become severely infectious agents; particularly they are frequently find novel mechanisms to evade the antibiotics treatments [1,2]. They developed multidrug resistance to different antibiotics in common use (MDR) with markedly increasing prevalence by contacting with contaminating surfaces and apparatus or infected persons [3]. They gain increasing concern due to their facility for withstanding the influence of various antibacterial agents, accordingly limit the drug of choices and leads to higher mortality and morbidity [4]. Therefore, finding of alternate powerful, inexpensive and harmless natural agents against multidrug resistant bacteria can be potent way for solving this serious global problem [5-7]. Marine bacteria are promising reservoirs of diverse effective bioactive natural products and many of them are being used in chemotherapy to treatment human diseases especially with the continuing need for new potent compounds against drug-resistant pathogens and managing of distressing cancers with high selective activity and less toxicity [8,9]. Red sea host diverse and abundant free living microorganisms have the ability to produce promising bioactive marine natural products [10]. Marine Bacillus species produce multipurpose biologically active compounds including lipopeptide, polypeptide, macrolactone, fatty acid, polyketide and isocoumarin metabolites [11,12] with wide variety of antifungal, antibacterial, antioxidant and antiproliferative activities [13,14]. The capability of Bacillus species to biosynthesis different antibiotics with varied structures has been demonstrated by numerous genetic studies, and genetic analysis of Bacillus strains has shown that about 8% of the genome is dedicated to antibiotic synthesis [15,16]. This work was aimed to assess the incidences and distributions of Enterococci among patient admitted to El-Demerdash teaching hospital (Cairo, Egypt) along with determining their antibiotic sensitivity profiles against a panel of antibiotics to select the multidrug resistant Enterococci (MDRE). Moreover, this study aimed to explore potent marine Bacillus species that able to produce biologically active substances against these clinical isolates and diverse malignant cell lines including colon, liver, breast and lung carcinomas.
Clinical specimens
Different samples including urine, pus, blood, wound, sputum and synovial fluid (5 samples for each) collected from patients ranging from 1 to over 60 years old (15 females and 15 males) admitted to I.C.U (intensive care unit), surgery, diabetic, skin and venereal disease, hematology, emergency, E.N.T (ear, nose and throat) and neurology units of El-Demerdash teaching hospital, Cairo, Egypt. Samples were brought to the laboratory under iced conditions and promptly processed. Fifty nine Enterococci isolates obtained were involved in this work. Moreover, Enterococcus faecalis ATCC 19433, Enterococcus faecium ATCC 19434, Enterococcus raffinosus ATCC 49427, Enterococcus durans ATCC 19432, Enterococcus mundtiiATCC 43186 and Enterococcus avium ATCC 14025 used as reference strains. Different samples including urine, pus, blood, wound, sputum and synovial fluid (5 samples for each) collected from patients ranging from 1 to over 60 years old (15 females and 15 males) admitted to I.C.U (intensive care unit), surgery, diabetic, skin and venereal disease, hematology, emergency, E.N.T (ear, nose and throat) and neurology units of El-Demerdash teaching hospital, Cairo, Egypt. Samples were brought to the laboratory under iced conditions and promptly processed. Fifty nine Enterococci isolates obtained were involved in this work. Moreover, Enterococcus faecalis ATCC 19433, Enterococcus faecium ATCC 19434, Enterococcus raffinosus ATCC 49427, Enterococcus durans ATCC 19432, Enterococcus mundtiiATCC 43186 and Enterococcus avium ATCC 14025 used as reference strains.
Isolation and identification of clinical bacteria
Enterococcus isolates were isolated and characterized according to the traditional methods and biochemical key were previously reported [17-23].
Antibiotics susceptibility profile of clinical Enterococcalisolates
Antibacterial susceptibility test of Enterococci species was done following disk diffusion technique using Muller Hinton agar (MH) based on World Health Organization [24] and adopted as consensual standard by the Clinical Laboratory Standards Institute [25,26] and European Committee on antimicrobial susceptibility testing [27] against a panel of thirteen antibiotics including penicillin 10 IU, ciprofloxacin 5 μg, streptomycin 10 μg, vancomycin 30 μg, gentamicin 10 µg, tetracycline 30 μg, kanamycin 30 µg, linezolid 10 µg, chloramphenicol 30 µg, teicoplanin 30 µg, nitrofurantoin 100 µg, erythromycin 15 μg and oxacillin 5 µg (Oxoid, Basingstoke, Hampshire, England in µg/disk).
Marine samples and isolation of marine Bacillus species
Ten samples of seawater from the fishing area Masturah, Red Sea, Jeddah, Saudi Arabia (latitude: 23°5031.4600N/longitude: 38°49017.5200E) were collected in August 2018 at different depths in sterile screw cap bottles under iced conditions. Collected samples were taken to the laboratory, gathered and processed instantly. The isolation medium and process were prepared and done following the method of Ivanova, et al. [28] by plating serial dilutions of water sample individually to Petri dishes of nutrient agar (NA) supplemented with 100 µg/mL nystatin and cycloheximide, incubated at 30 °C for 3 days and recognized bacterial single colonies were transferred periodically to NA at 30 °C for 48 h and included in this study. Bacterial isolates were preserved on NA at 4 °C till using.
Antibacterial activity of marine bacterial isolates against different MDR-Enterococci
Muller Hinton agar (MH) plates were inoculated with the clinical MDR-Enterococci isolates, individually and paper assay discs loaded with 30 μL of marine bacterial isolates supernatants separately were plated on the top of inoculated medium, incubated at 37 °C for 24 h and then antimicrobial activity of bacterial isolates was determined against the MDRE isolates by using the routine diffusion plate technique via evaluating the inhibition zone diameters in mm [data determined as no antagonistic activity (-),weak antagonistic activity (˂10 mm, +), moderate antagonistic activity (10 - 15 mm, ++) and excellent antagonistic activity (16 - ˃20 mm, +++)] [11,29].
Phenotypic and chemotypic properties of marine bacterial isolate ESRAA3010
ESRAA3010 strain was specified by conventional taxonomic procedures by means of API 20E and API 50CH methods along with other phenotypic and chemotypic characters [29-32]. Bacillus subtilis ATCC 6051T used as standard strain.
Molecular identification of isolate ESRAA3010 by 16S rDNA sequence analysis
DNA extraction, PCR amplification of 16S ribosomal RNA (rRNA) gene, purification of the PCR products, gel electrophoreses, and the 16S rDNA sequence analysis were performed based on previous reports [8,31,33-35] followed by aligning the 16S rRNA gene sequence of isolate ESRAA3010 with published sequences in NCBI GenBank database (http://www.ncbi.nih.gov). The tree topology was assessed through neighbor-joining method and bootstrap analyses based on 500 replications with MEGA-X [36-38].
Extraction of bioactive metabolites from marine B. subtilis ESRAA3010
Bacillus subtilis ESRAA3010 was inoculated into Erlenmeyer flasks containing tryptic soy broth medium then incubated for 24 h at 30 °C and 100 rpm, after incubation period the fermented broth of ESRAA3010 strain (5 L) was collected and the supernatant was separated under reduced pressure and then extracted twice with ethyl acetate (1:1, pH 4.5 under overnight shaking). The EtOAc extract obtained evaporated to dryness giving light brownish oil (6.41 g).
Determination of minimum inhibitory (MIC) and minimum bactericidal concentrations (MBC) of B. subtilis ESRAA3010 extract against Enterococci strains.
The MIC and MBC of the extracted secondary metabolites were estimated in µg/mL against MDRE isolates as described by Cappuccino and Sherman [39] and Lavermicocca, et al. [40].
Determination of anti-proliferative activity (MTT assay) of the Bacillus subtilis ESRAA3010 on the colon (HCT- 116), liver (HepG-2), Breast (MCF-7) and lung (A-549) carcinomas.
Cell viability test was measured by the mitochondrial dependent reduction of yellow MTT (3-(4, 5-dimethylthiazol-2-yl)-2, 5- diphenyl tetrazolium bromide) to purple formazan [41,42]. HCT-116, MCF-7, HepG-2 and A-549 were achieved from Cancer Center, Karolinska Institute, Stokholm, Sweden; DMEM medium, RPMI 1640 medium and 1% antibiotic-antimycotic (10,000 U/mL potassium penicillin; 10,000 μg/mL streptomycin sulfate and 25 μg/mL amphotericin B) were achieved from Life Technologies/Gibco (Grand Island, NY, USA). The effect of diverse amounts of B. subtilis ESRA-3010 extract ranging between 25 and 300 µg/mL on the cytotoxicity and cell viability of these tumor cell lines was measured as stated by Mosmann [41] and Wilson [42] in 96-well microplate at 37 °C under 5% CO2 using a water jacketed CO2 incubator (Sheldon, TC2323, Cornelius, OR, USA) for 48 h followed by aspirating medium, adding 40 μL MTT salt (2.5 μg/mL) to wells, incubating for 4 h at 37 °C as mentioned above, ending the reaction along with dissolve the resulted crystals by adding 200 μL of 10% sodium dodecyl sulfate to wells and thus incubating overnight at 37 °C. The absorbance was estimated at 595 nm by the microplate multi-well reader (Bio-Rad Laboratories Inc. model 3350, Hercules, California, USA). The IC50 (the amount of the extract that decreased cell viability by 50%) compared to the control (wells contain only solvent without any extract) were determined using SPSS 11 program. Percentages (%) of cell viabilities were estimated based on the formula: [(Absorbance of treated cell lines with extract / Absorbance of negative control) -1] X 100.
Isolation, characterization and occurrence of clinical Enterococci isolates
A total of fifty nine isolates of Gram-positive Enterococci isolates including 16, 11, 8, 5, 10 and 9 isolates were isolated from the age groups 1-6, ˃6-12, ˃12-20, ˃20-40, ˃40-60 and over 60 years old (Table 1). The largest number of MDRE (27.12%) was collected from ages ranging from 1 to 6 years. Furthermore, 33 isolates (55.93%) were obtained from male samples while 26 isolates (44.07%) were obtained from female participants (Table 1) from El-Demerdash teaching hospital. The presence of resistant Enterococci isolates in human societies acts as a source for hospital infections [1]. In line with our results, Karna, et al. [4] revealed that the predominance of Enterococci isolates were achieved from age group 0-10 (20.9%), after that age group 20-30 (19.8%) along with detection of the high incidence of infection in male participants. Enterococci are recognized as unique reasons of hospital infections in patients with weakened immune systems. On the other hand, Table 2 showed the different department from which specimens were collected, 17 (28.81%); 16 (27.12%); 8 (13.56%); 7 (11.86%); 5 (8.48%); 4 (6.78%); 1 (1.70%) and 1 (1.70%) isolates were obtained from I.C.U (intensive care unit), surgery, diabetic, skin and venereal disease, hematology, emergency, E.N.T (ear, nose and throat) and neurology departments, respectively which indicating that hospitalization in the intensive care unit is an important risk factor for MDRE colonization and occurrence. Similar distribution observation were reported previously in resistant Enterococci collated from different humanoid infections [19], clinical samples in Kashmir; North India [43], health care setting [3], tertiary care center of Eastern Nepal [4] and Turkey [1].
| Table 1: Specimens recovered from different gender and age groups. | ||
| Demographic and clinical conditions | No. of clinical isolates | % of clinical isolates |
| Gender | ||
| Males | 33 | 55.93 |
| Females | 26 | 44.07 |
| Age | ||
| 1-6 | 16 | 27.12 |
| >6-12 | 11 | 18.64 |
| >12-20 | 8 | 13.56 |
| >20-40 | 5 | 8.48 |
| >40-60 | 10 | 16.95 |
| >60 | 9 | 15.25 |
| Table 2: Incidence of Enterococci isolates in various departments at El-Demerdash teaching hospital. | ||
| Department | No. of Enterococci isolates | % of Enterococci isolates |
| Surgery | 16 | 27.12 |
| I.C.U. | 17 | 28.81 |
| Diabetic | 8 | 13.56 |
| Emergency | 4 | 6.78 |
| E.N.T. | 1 | 1.7 |
| Neurology | 1 | 1.7 |
| Haematology | 5 | 8.48 |
| Skin and venereal disease | 7 | 11.86 |
| Total | 59 | 100 |
Characteristics of clinical Enterococcus strains
Following the standard guideline of species specification [2,17-20], six different species of Enterococci isolates were identified, namely E. faecalis, E. faecium, E. raffinosus, E. durans, E. mundtii and E. avium (n = 35, 15, 4, 3, 1 and 1, respectively, Table 3. All isolates were Gram-positive, non-motile, positive for Voges-Proskauer reaction, negative for catalase activity and β-glucoronidase, produce acid from sorbitol and lactose as well as grown at 45 °C and pH 9.6. Only E. mundtii was able to produce yellow pigment which is a key feature of this species in addition to produce acid from trehalose; E. durans was the only negative strain for acid production from rhamnose, melezitose, arabinose and mannitol and E. avium was the only strain failed to produce acid from amidon but gelatin hydrolysis ability was only recorded in E. faecalis strains (Table 3). The other details of phenotypic characterizations and chemotypic features of these species obtained in the current study are presented in table 3. Interestingly E. faecalis and E. faecium (Figures 1 and 2) together constitute more than 84.74% of total isolates and their increased proportion in current work might be attributed to their capability to attain and developed different resistance patterns against multiple antibiotics. In the majority of previous reports, E. faecalis has been documented as the main Enterococci species, followed by E. faecium. For example Karna, et al. [4] reported that among seven different identified species of Enterococci the highest frequency of strains among total isolates was reported for E. faecalis followed by E. faecium and together, they made up over 90% of the total isolates. Nevertheless, this result is higher than other studies [44,45]. These differences in bacterial occurrence can be attributed to the differences in geographic site, sample, period of hospitalization, and drugs used [46].
| Table 3: Characteristics of clinical Enterococcus strains. | ||||||
| Characteristic | No. of positive Enterococci strains | |||||
| E. faecalis n = 35 |
E. faecium n = 15 |
E. raffinosus n = 4 |
E. durans n = 3 |
E. mundtii n = 1 |
E. avium n = 1 |
|
| Gram stain | 35 | 15 | 4 | 3 | 1 | 1 |
| Motility | 0 | 0 | 0 | 0 | 0 | 0 |
| Pigment (yellow) | 0 | 0 | 0 | 0 | 1 | 0 |
| Voges-Proskauer reaction | 35 | 15 | 4 | 3 | 1 | 1 |
| Hippurate hydrolysis | 29 | 12 | 3 | 0 | 0 | 0 |
| α-Galactosidase | 0 | 15 | 2 | 0 | 1 | 0 |
| β-Galactosidase | 11 | 14 | 0 | 0 | 1 | 0 |
| Arginine dihydrolase | 35 | 15 | 0 | 3 | 1 | 0 |
| β-Glucuronidase | 0 | 0 | 0 | 0 | 0 | 0 |
| Acid from | ||||||
| Amidon | 35 | 15 | 3 | 3 | 1 | 0 |
| Glycogen | 7 | 0 | 0 | 0 | 1 | 0 |
| Sucrose | 31 | 14 | 2 | 0 | 0 | 1 |
| Sorbose | 13 | 0 | 0 | 0 | 0 | 1 |
| Rhamnose | 32 | 3 | 1 | 0 | 1 | 1 |
| Melibiose | 9 | 14 | 4 | 0 | 1 | 0 |
| Melezitose | 0 | 13 | 4 | 0 | 1 | 1 |
| L-Arabinose | 33 | 12 | 4 | 0 | 1 | 1 |
| Mannitol | 34 | 0 | 4 | 0 | 1 | 1 |
| Sorbitol | 35 | 15 | 3 | 3 | 1 | 1 |
| Lactose | 35 | 15 | 4 | 3 | 1 | 1 |
| Trehalose | 0 | 0 | 0 | 0 | 1 | 0 |
| Inulin | 0 | 15 | 0 | 0 | 1 | 0 |
| Raffinose | 30 | 1 | 1 | 0 | 0 | 1 |
| D-xylose | 25 | 5 | 1 | 0 | 1 | 0 |
| Adonitol | 0 | 0 | 2 | 0 | 0 | 1 |
| Gelatinase production | 24 | 0 | 0 | 0 | 0 | 0 |
| H2S production | 0 | 0 | 0 | 0 | 1 | 1 |
| β-hemolysis | 10 | 3 | 0 | 2 | 0 | 0 |
| Catalase | 0 | 0 | 0 | 0 | 0 | 0 |
| Growth at | ||||||
| 4 °C | 0 | 15 | 2 | 1 | 1 | 0 |
| 45 °C | 35 | 15 | 4 | 3 | 1 | 1 |
| 50 °C | 0 | 11 | 4 | 0 | 1 | 0 |
| pH 9.6 | 35 | 15 | 4 | 3 | 1 | 1 |
| 0.01% Tetrazolium | 35 | 0 | 2 | 0 | 0 | 0 |
Figure 1: E. faecalis (a) and E. faecium (b) under microscope.
Figure 2: Appearance of E. faecalis (a) and E. faecium (b) on MacConkey agar.
Enterococci species distribution in various clinical specimens
Among a total number of 59 Enterococci isolates obtained, 12 (20.34%), 20 (33.9%), 9 (15.25%), 10 (16.95%), 6 (10.17%) and 2 (3.39%) were recovered from urine, pus, blood, wound, sputum and synovial fluid samples, respectively (Table 4). After analyzing the distribution of the six Enterococci species in the various clinical specimens obtained, we found that 8 (13.56%), 11 (18.64%), 6 (10.17%), 5 (8.48%), 3 (5.09%) and 2 (3.39%) of E. faecalis (n = 35, 59.32%) as well as 3 (5.09%), 5 (8.48%), 3 (5.097%), 3 (5.097%), 1 (1.70%) and 0 (0.00%) isolates of E. faecium (n = 15, 25.42%) along with 1 (1.70%), 2 (3.39%), 0 (0.00%), 1 (1.70%), 0 (0.00%) and 0 (0.00%) of E. raffinosus (n = 4, 6.78%) were obtained from urine, pus, blood, wound, sputum and synovial fluid samples, respectively. Moreover only 3 isolates of E. durans (1 isolate from pus and 2 isolates from sputum), E. mundtii (1 isolate from pus) and E. avium (1 isolate from wound) were isolated (Table 4). In agreement with our results, Karna, et al. [4] analyzed ninety one isolates of Enterococcus obtained from numerous clinical samples, among them the highest Enterococci isolates incidence reported from urine then pus and blood (61.5%, 19.8% and 5.5%, respectively).
| Table 4: Distributionof Enterococcus species in the clinical specimens. | |||||||
| Specimen | Total no. of Enterococcus n, (%) | Enterococci species n, (%) | |||||
| E. faecalis | E. faecium | E. raffinosus | E. durans | E. mundtii | E. avium | ||
| Urine | 12 (20.34) | 8 (13.56) | 3 (5.09) | 1 (1.70) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Pus | 20 (33.90) | 11 (18.64) | 5 (8.48) | 2 (3.39) | 1 (1.70) | 1 (1.70) | 0 (0.00) |
| Blood | 9 (15.25) | 6 (10.17) | 3 (5.09) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Wound | 10 (16.95) | 5 (8.48) | 3 (5.09) | 1 (1.70) | 0 (0.00) | 0 (0.00) | 1 (1.70) |
| Sputum | 6 (10.17) | 3 (5.09) | 1 (1.70) | 0 (0.00) | 2 (3.39) | 0 (0.00) | 0 (0.00) |
| Synovial fluid | 2 (3.39) | 2 (3.39) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Total | 59 (100.00) | 35 (59.32) | 15 (25.42) | 4 (6.78) | 3 (5.09) | 1 (1.70) | 1 (1.70) |
Antibiotic susceptibility pattern in the Enterococci isolates
In the current work, the susceptibility patterns in the Enterococci isolates against antibiotics in common use by the disc diffusion method in table 5 displayed that all Enterococci isolates were multidrug resistant (MDRE) due to they showed co-resistant to many classes of antibiotics at the same time. The multidrug resistant (MDR) considered particular phenotypic characteristics of the clinical Enterococci strains [1,47]. The highest resistant was reported for penicillin (n = 53, 89.83%), oxacillin (n = 50, 84.75%), erythromycin (n = 49, 83.05%), streptomycin (n = 47, 79.66%), ciprofloxacin (n = 42, 71.19%), kanamycin (n = 41, 69.49%), tetracycline (n = 40, 67.80%), gentamicin (n = 33, 55.93%), chloramphenicol (n = 31, 52.54%) and nitrofurantoin (n = 29, 49.15%) but the lowest resistant was detected with linezolid (n = 5, 8.48%), followed by teicoplanin (n = 14, 23.73%) and vancomycin (n = 20, 33.90%) (Table 5). Karna, et al. [4] stated that highest frequency of susceptibility among the isolates of Enterococci was noted for linezolid after that teicoplanin and then gentamicin (97.8%, 95.6% and 81.3%, respectively). Interestingly 94.29%, 88.57%, 42.86%, 85.71%, 80.0%, 31.43%, 77.14%, 100.00%, 11.43%, 25.71%, 57.14%, 85.71% and 94.29% of total E. faecalis isolates compared to 93.33%, 80.00%, 20.00%, 20.00%, 33.33%, 60.00%, 66.70%, 86.67%, 6.67%, 26.66%, 46.67%, 53.33% and 86.67% of total E. faecium isolates showed resistant to penicillin, streptomycin, vancomycin, gentamicin, tetracycline, nivtrofurantoin, ciprofloxacin, erythromycin, linezolid, teicoplanin, chloramphenicol, kanamycin and oxacillin, respectively but E. raffinosus, E. durans, E. mundtii and E. avium species showed less degree of resistant to these antibiotics (Table 5). The higher positive resistance rate to all antibiotic under study demonstrated in our work agreed well with other studies [2,3,20] that could be attributed to the low affinity between these antibiotics and protein binding sites of E. faecium, E. raffinosus, E. durans, E. mundtii and E. avium or/and the presence of plasmid-encoded β-lactamase and other antibiotics degrading enzymes in some strains than others [6,48]. Our findings are in agreement with many studies reported E. faecalis as the most frequently species obtained from hospitalized patient with multidrug resistance against different antibiotics including vancomycin [44,49]. Conversely Karna, et al. [4] suggested that E. faecium strains were higher resistant to all antibiotics under study than E. faecalis but E. durans showed no resistant to any of the tested antibiotics.
| Table 5: Antibiotic resistant profiles of the Enterococcus strains. | ||||||||||
| Antibiotics | Enterococcus strains | |||||||||
| Total Enterococci | E. faecalis (n = 35) | E. faecium (n = 15) | Other Enterococci (n = 9) | |||||||
| No. of total resist Enterococci | Percentage of resistance | No. of resist isolates | % of resistance* | No. of resist isolates | % of resistance* | No. of resist isolates | % of resistance* | |||
| A | B | A | C | D | ||||||
| Penicillin | 53 | 89.83 | 33 | 55.93 | 94.29 | 14 | 23.73 | 93.33 | 6 | 66.67 |
| Streptomycin | 47 | 79.66 | 31 | 52.54 | 88.57 | 12 | 16.95 | 80 | 4 | 44.44 |
| Vancomycin | 20 | 33.9 | 15 | 25.42 | 42.86 | 3 | 5.09 | 20 | 2 | 22.22 |
| Gentamicin | 33 | 55.93 | 30 | 50.85 | 85.71 | 3 | 5.09 | 20 | 0 | 0 |
| Tetracycline | 40 | 67.8 | 28 | 47.46 | 80 | 5 | 8.48 | 33.33 | 7 | 77.8 |
| Nitrofurantoin | 29 | 49.15 | 11 | 18.64 | 31.43 | 9 | 15.25 | 60 | 9 | 100 |
| Ciprofloxacin | 42 | 71.19 | 27 | 45.76 | 77.14 | 10 | 16.95 | 66.7 | 5 | 55.6 |
| Erythromycin | 49 | 83.05 | 35 | 59.32 | 100 | 13 | 22.03 | 86.67 | 1 | 11.11 |
| Linezolid | 5 | 8.48 | 4 | 6.78 | 11.43 | 1 | 1.7 | 6.67 | 0 | 0 |
| Teicoplanin | 14 | 23.73 | 9 | 15.25 | 25.71 | 4 | 4.21 | 26.66 | 1 | 11.11 |
| Chloramphenicol | 31 | 52.54 | 20 | 33.9 | 57.14 | 7 | 11.87 | 46.67 | 4 | 44.44 |
| Kanamycin | 41 | 69.49 | 30 | 50.85 | 85.71 | 8 | 13.56 | 53.33 | 3 | 33.33 |
| Oxacillin | 50 | 84.75 | 33 | 55.93 | 94.29 | 13 | 22.03 | 86.67 | 4 | 44.44 |
| *A = % of total Enterococci, B = % of total E .faecalis, C = % of total E. faecium, D = % of totalother Enterococci (E. raffinosus, E. durans, E. mundtii and E. avium). | ||||||||||
Isolation and evaluation of antagonistic activity of different marine Bacillus isolates against different Enterococci strains
Twelve isolates of free living marine Bacillus species were isolated, cultivated, and then their antagonistic activity toward Enterococci strains under study was evaluated and tabulated in table 6. Data clearly indicated that the isolate under the isolation code ESRAA3010 was the hyperactive strain that showed inhibitory activity against all Enterococci strains under study with inhibition power ranged from good (++) to excellent (+++) followed by ESRAA3012 isolate showed inhibitory activity toward 93.22% of all isolates (Table 6). Then ESRAA3010 strain was selected for the further studies. Our results supported the previous finding of Lv, et al. Mondol, et al., and Freitas-Silva, et al. [13,14,50] they documented that marine Bacillus strains can biosynthesis multipurpose compounds comprising lipopeptide, carotenoid, polypeptide, macrolactone, fatty acid, polyketide and isocoumarin metabolites that have demonstrated a wide array of bioactivities including antibacterial, antifungal, antitumor and antioxidant properties. Consequently, there is a potential for using these marine Bacillus species metabolites as promising medicines and in other biological treatments.
| Table 6: Antagonistic activity of marine Bacillus isolates against clinical Enterococcus strains. | ||||||
| Antagonistic marine Bacillus isolates | No. of sensitive Enterococci strains / inhibition activity range* | |||||
| E. faecalis n = 35 | E. faecium n = 15 | E. raffinosus n = 4 | E. durans n = 3 | E. mundtii n = 1 | E. avium n = 1 | |
| Bacillus sp. ESRAA3001 | 20 (+ to +++) | 9 (+ to ++) | 3 (+ to ++) | (-) | (-) | (-) |
| Bacillus sp. ESRAA3002 | (-) | (-) | (-) | 1 (++) | 1 (+) | 1 (+) |
| Bacillus sp. ESRAA3003 | 4 (+ to ++) | 6 (+ to ++) | 3 (+ to +++) | (-) | (-) | 1 (++) |
| Bacillus sp. ESRAA3004 | 11 (+ to ++) | 6 (+ to ++) | 3 (+) | 3 (+ to ++) | (-) | (-) |
| Bacillus sp. ESRAA3005 | 9 (+ to +++) | 5 (++) | (-) | 2 (++) | 1 (+) | 1 (++) |
| Bacillus sp. ESRAA3006 | 25 (+ to ++) | 13 (++) | 1 (+++) | (-) | (-) | (-) |
| Bacillus sp. ESRAA3007 | 18 (+ to +++) | 14 (++ to +++) | (-) | 1 (++) | 1 (++) | 1 (++) |
| Bacillus sp. ESRAA3008 | (-) | (-) | (-) | (-) | (-) | (-) |
| Bacillus sp. ESRAA3009 | (-) | (-) | (-) | (-) | (-) | (-) |
| Bacillus sp. ESRAA3010 | 35 (++ to +++) | 15 (+++) | 4 (+++) | 3 (+++) | 1 (+++) | 1 (+++) |
| Bacillus sp. ESRAA3011 | (-) | (-) | (-) | (-) | (-) | (-) |
| Bacillus sp. ESRAA3012 | 35 (+ to +++) | 15 (+ to ++) | 3 (+++) | 1 (+++) | 1 (+++) | (-) |
| * (-) no inhibitory activity, (+) weak inhibitory activity, moderate inhibitory activity (++) and (+++) excellent inhibitory activity | ||||||
Identification of the hyperactive bacteria ESRAA3010 by phenotypic, chemotypic and 16S rDNA analysis
Strain ESRAA3010 showed a grayish-white, roundish, opaque, flatted and medium size colonies ranged from drying on LB agar and nutrient agar to smooth and moist on tryptic soy agar in addition to complete hemolytic activity on blood agar (Table 7). It was Gram positive rods, spore forming, cell diameter estimated to be 0.8–0.9 and 2.7–3.2 μm in width and length, respectively (Figure 3). It was positive for oxidase, catalase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, β-fucosidase, α-arabinosidase, L-arginine aminopeptidase, VP reaction and nitrate reduction; acid production form D-glucose, L-arabinose, D-xylose, D-mannitol and D-frucrose; citrate utilization; hydrolysis of starch, gelatin, casein and tween 80 as well as assimilation of L-arabinose, D-xylose, meso-Inositol, sorbitol, methyl-D-glucoside, D-melibiose, D-raffinose and glycogen (Table 7).
Figure 3: Gram staining of isolate ESRAA3010 under microscope.
| Table 7: Identification and characterization of marine isolate ESRAA3010. | |
| Physiological and biochemical test | Characteristics |
| Growth performance on | |
| TSA agar + 5% fetal calf serum | Grayish white, roundish, opaque, thick ridges, smooth, moist and medium size colonies |
| LB agar | Grayish white, roundish, opaque, flat, dry and medium size colonies |
| Rabbit blood agar | Grayish white, roundish, complete hemolysis, opaque, flat, dry and medium size colonies |
| Nutrient agar | Grayish white, roundish, opaque, flat, dry and medium size colonies |
| Shape | Rod |
| Gram stain | Positive |
| Cell diameter | 0.8–0.9 X 2.7–3.2 μm |
| Indol reaction | - |
| Aminopeptidase | - |
| KOH test | - |
| Oxidase | + |
| Catalase | + |
| Phenylalanine deaminase | - |
| Arginine dihydrolase | - |
| α-Galactosidase | + |
| β- Galactosidase | + |
| α- Glucosidase | + |
| β-Glucosidase | + |
| β-Fucosidase | + |
| α-Arabinosidase | + |
| L-Arginine aminopeptidase | + |
| N-benzoyl-L-leucine amino peptidase | - |
| L-Tryptophan aminopeptidase | - |
| Spores | + |
| VP reaction | + |
| Growth at | |
| 45 °C | + |
| 50 °C | - |
| 60 °C | + |
| pH 5.5 | + |
| NaCl 5% | + |
| NaCl 10% | + |
| NaCl 15% | + |
| Acid form | |
| D-Glucose | + |
| L-Arabinose | + |
| D-Xylose | + |
| D-Mannitol | + |
| D-Frucrose | + |
| Utilization of | |
| Citrate | + |
| Propionate | - |
| Nitrate | + |
| Hydrolysis of | |
| Starch | + |
| Gelatin | + |
| Casein | + |
| Tween 80 | + |
| Assimilation of | |
| L-Arabinose | + |
| D-Xylose | + |
| Galactose | - |
| Rhamnose | - |
| meso-Inositol | + |
| Sorbitol | + |
| Methyl-D-mannoside | - |
| Methyl-D-glucoside | + |
| N-acetyl-glucosamine | - |
| D-Melibiose | + |
| D-Melibios | + |
| D -Raffinose | + |
| Dextrin | - |
| Starch | + |
| Glycogen | + |
| Gluconate | - |
On the other hand, it was negative for KOH test, indol reaction, aminopeptidase, phenylalanine deaminase, arginine dihydrolase, N-benzoyl-L-leucine amino peptidase and L-tryptophan aminopeptidase; utilization of propionate and assimilation of galactose, rhamnose, methyl-D-mannoside, N-acetyl-glucosamine, dextrin and gluconate (Table 7).
In addition, the sequencing of the 16S rRNA gene exhibited 99.93% similarity to various B. subtilis strains (DSM 10, JCM 1465 and NBRC 13719). The phenotypic, chemotaxonomic, 16S rRNA gene analyses and phylogenetic data in table 7 and figures 3,4 showed that isolate ESRAA3010 is very close to B. subtilis as previously reported [8,29-35]. Then it was designated as B. subtilis ESRAA3010.
Figure 4: Phylogenetic dendrogram of isolate ESRAA3010 based on 16S rDNA sequence analysis, constructed using the neighbor-joining method.
Minimum inhibitory (MIC) and minimum bactericidal concentrations (MBC) of B. subtilis ESRAA3010 extract against different Enterococci strains
Data in table 8 indicated that EtOAc extract of B. subtilis ESRAA3010 showed potent antagonistic activity against all Enterococci species under study. It showed MIC against E. faecalis, E. faecium, E. raffinosus, E. durans, E. mundtii and E. avium (35, 15, 4, 3, 1 and 1isolates, respectively) ranged from 4 to 56 µg/mL, 4 to 12 µg/mL, 4 to 8 µg/mL, 4 to 8 µg/mL, 8 µg/mL and 4 µg/mL, respectively and MBC reached 8 to 64 µg/mL, 4 to 16 µg/mL, 4 to 12 µg/mL, 4 to 16 µg/mL, 12 µg/mL and 8 µg/mL, respectively. Our data are consistent with Kizhakkekalam, et al. and Freitas-Silva, et al. [11,50] they demonstrated the antagonistic activity of secondary metabolites derived from marine Bacillus species as potential antimicrobial agents against multidrug-resistant bacteria.
Anti-proliferative activity of B. subtilis ESRAA3010 extract
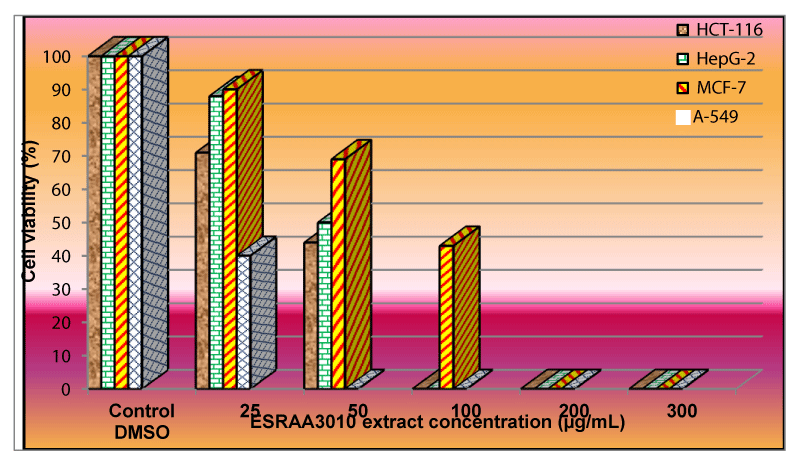
In vitro anti-proliferative activity of EtOAc extract of B. subtilis ESRAA3010 strain on different tumor cell lines was evaluated by MTT cell viability assay and illustrated in figure 5. Data clearly indicated that the cells viability of A-549 carcinoma was completely inhibited after treatment with ESRAA3010 extract at a concentration of 50 µg/mL (Figure 5). Furthermore, HCT-116 and HepG-2 carcinomas totally inhibited at 100 µg/mL but the growth of MCF-7 cell line was completely inhibited at a concentration of 200 µg/mL with IC50 equal to 39, 50, 75 and 19 µg/mL against colorectal (HCT-116), hepatocellular (HepG-2), breast (MCF-7) and lung (A-549) carcinomas. Similar to our results Vo, et al., Koim-Puchowska, et al. and Mondol, et al. [7,12,14] reported the anticancer activities of Bacillus species extracts on a large number of carcinomas. Also, Zhao, et al. [16] reported that treating carcinomas with products from cultured Bacillus strains had significant inhibitory effects on ovarian and colorectal carcinomas proliferation in a dose dependent manner.
Figure 5: Effect of B. subtilis ESRAA3010 extract against HCT-116, HepG-2, MCF-7 and A-549 carcinomas.
Marine ecosystems in Egypt have proven to be prolific resource for various types of marine bacteria, especially Bacillus species that produce different stimulating biological metabolites against infectious agents which widely distributed as multidrug resistant Enterococci strains and cancer. The ethyl acetate extract of B. subtilis ESRAA3010 showed potent anti-MDRE activity against all clinical Enterococci isolates under study including E. faecalis, E. faecium, E. raffinosus, E. durans, E. mundtii and E. avium with MIC 4 to 56, 4 to 12, 4 to 8, 4 to 8, 8 and 4 µg/mL, respectively and MBC 8 to 64, 4 to 16, 4 to 12, 4 to 16, 20 and 8 µg/mL, respectively. Bacillus subtilis ESRAA3010 extract exhibited anti-proliferative activity against colon, lung, liver and breast adenocarcinomas with IC50 equal to 39, 50, 75 and 19 µg/mL, respectively. Our data supported the potential application of B. subtilis ESRAA3010 extract in drug delivery and industry as novel and promising antibacterial and/or anticancer agents against deadly infectious agents and cancers.
- Asgin N, Otlu B. Antibiotic resistance and molecular epidemiology of vancomycin-resistant Enterococci in a tertiary care hospital in Turkey. Infect Drug Resist. 2020; 13: 191-198. PubMed: https://pubmed.ncbi.nlm.nih.gov/32021333/
- Jaiswal S, Singh A, Verma RK, Singh DP, Kumari S. Characterization, speciation and antimicrobial resistance pattern of Enterococcus species isolated from clinical specimens at a rural tertiary care hospital. Int J Res Med Sci. 2017; 5: 3484-3487.
- Faron ML, Ledeboer NA, Buchan BW. Resistance mechanisms, epidemiology and approaches to screening for vancomycin-resistant Enterococcus in the health care setting. J Clin Microbiol. 2016; 54: 2436-2447. PubMed: https://pubmed.ncbi.nlm.nih.gov/27147728/
- Karna A, Baral R, Khana B. Characterization of clinical isolates of Enterococci with special reference to glycopeptide susceptibility at a tertiary care center of Eastern Nepal. Int J Microbiol. 2019; 2019: 7936156.
- El-Gendy MMA, Mohamed ZK, Hekal NZ, Ali FM, Yousef AEM. Production of bioactive metabolites from different marine endophytic Streptomyces species and testing them against methicillin-resistant Staphylococcus aureus (MRSA) and cancer cell lines. BioTechnologia. 2018a; 99: 13-35.
- Nasaj M, Mousavi SM, Hosseini SM, Arabestani MR. Prevalence of virulence factors and vancomycin-resistant genes among Enterococcus faecalis and E. faecium isolated from clinical specimens. Iran J Public Health 2016; 45: 806-813. PubMed: https://pubmed.ncbi.nlm.nih.gov/27648425/
- Vo TTT, Lee CW, Wu CZ, Liu JF, Lin WN, et al. Surfactin from Bacillus subtilis attenuates ambient air particulate matter-promoted human oral cancer cells metastatic potential. J Cancer. 2020; 11: 6038-6049.
- Ki JS, Zhang W, Qia PY. Discovery of marine Bacillus species by 16S rRNA and rpoB comparisons and their usefulness for species identification. J Microbiol Methods. 2009; 77: 48-57. PubMed: https://pubmed.ncbi.nlm.nih.gov/19166882/ .
- Viju N, Punitha SMJ, Satheesh S. Antibiofilm activity of symbiotic Bacillus species associated with marine gastropods. Ann Microbiol. 2020; 70: 11.
- El-Gendy MMA, Abdel-Wahhab KG, Mannaa FA, Farghaly AA, El- Bondkly AMA. Carcinogenic activities and sperm abnormalities of methicillin resistance Staphylococcus aureus and inhibition of their virulence potentials by ayamycin. Appl Biochem Biotechno. 2017; 183: 833-852. PubMed: https://pubmed.ncbi.nlm.nih.gov/28389766/
- Kizhakkekalam VK, Chakraborty K, Joy M. Oxygenated elansolid-type of polyketide spanned macrolides from a marine heterotrophic Bacillus as prospective antimicrobial agents against multidrug-resistant pathogens. Int J Antimicrob Agents. 2020; 55: 105892.
- Koim-Puchowska B, Kłosowski G, Mikulski D, Menka A. Evaluation of various methods of selection of B. subtilis strains capable of secreting surface-active compounds. PLoS One. 2019; 14: e0225108. PubMed: https://pubmed.ncbi.nlm.nih.gov/31715626/
- Lv J, Da R, Cheng Y, Tuo X, Wei J, et al. Mechanism of antibacterial activity of Bacillus amyloliquefaciens C-1 lipopeptide toward anaerobic Clostridium difficile. Biomed Res Int. 2020; 2020: 3104613. PubMed: https://pubmed.ncbi.nlm.nih.gov/32190658/
- Mondol MAM, Hee JS, Mohammad TI. Diversity of secondary metabolites from marine Bacillus species: chemistry and biological activity. Mar Drugs. 2013; 11: 2846-2872.
- Liu Y, Lai Q, Du J, et al. Genetic diversity and population structure of the Bacillus cereus group bacteria from diverse marine environments. Sci Rep. 2017; 7: 689.
- Zhao M, Liang G, Zhou Y, et al. Novel Bacillus strains from the human gut exert anticancer effects on a broad range of malignancy types. Invest New Drugs. 2020; 38: 1373-1382.
- Collins MD, Jones D, Farrow JAE, Kilpper-Balz R, Schleifer KH. Enterococcus avium nom. rev., comb. nov.; E. cusselifluvus nom. rev., comb. nov.; E. durans nom. rev., comb. nov.; E. gallinarum comb. nov.; and E. malodoratus sp. nov. Int J Syst Bacteriol 1984; 34: 220-223.
- Devriese LA, Van De Kerckhove A, Kilpper-Balz R, Schleifer KH. Characterization and identification of Enterococcus species isolated from the intestines of animals. Int J System Bacteriol. 1987; 1987: 257-259.
- Facklam RR, Collins MD. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989; 27: 731-734. PubMed: https://pubmed.ncbi.nlm.nih.gov/2656745/
- Haritsa KB, Shashikala N, Kumar SDC, Sampath S. Isolation, identification and speciation of Enterococci and their antimicrobial susceptibility in a tertiary care hospital. J Evol Med Dent Sci 2014; 3: 13893-13899.
- Manero A, Blanch AR. Identification of Enterococcus Spp. with a biochemical key. Appl Environ Microbiol. 1999; 65: 4425-4430.
- Winn WC. Koneman’s color atlas and textbook of diagnostic microbiology. Lippincott Williams & Wilkins. 2006; Philadelphia, PA, USA.
- Yilema A, Moges F, Tadele S, Endris M, Kassu A, et al. Isolation of Enterococci, their antimicrobial susceptibility patterns and associated factors among patients attending at the university of Gondar Teaching Hospital. BMC Infect Dis. 2017; 17: 276. PubMed: https://pubmed.ncbi.nlm.nih.gov/28412932/
- World Health Organization Expert Committee on biological standardization. Technical report series. 1992; 822. WHO, Geneva.
- CLSI: Clinical and Laboratory Standards Institute. Approved standard M2-A10. Performance standards for antimicrobial susceptibility tests. 2010a; 10th ed. CLSI, Wayne, Pa.
- CLSI: Clinical and Laboratory Standards Institute. CLSI document M100-S19. Performance standards for antimicrobial susceptibility testing. 2010b; 20th informational supplement, Wayne, Pa.
- EUCAST: European Committee on Antimicrobial Susceptibility Testing. EUCAST QC Tables V1.2. 2010.
- Ivanova EP, Mikhailov VV, Andreev LA. Marine bacilli and some approaches to their identification. Mikrobiol Zhurnal. 1992; 54: 27-33.
- Ivanova EP, Nicolau DV, Yumoto N, Taguchi T, Okamoto K, et aL. Impact of the conditions of cultivation and adsorption on antimicrobial activity of marine bacteria. Mar Biol. 1998; 130: 545-551.
- Donnel AGO, Norris JR, Berkeley RCW, Claus D, Kanek T, et al. Characterization of Bacillus subtilis, Bacillus pumilus, Bacillus licheniformis, and Bacillus amyloliquefaciens by pyrolysis gas-liquid chromatography, deoxyribonucleic acid-deoxyribonucleic acid hybridization, biochemical tests, and API systems. Int J System Bacteriol. 1980; 30: 448-459.
- Farrow JAE, Wallbanks S, Collins MD. Phylogenetic interrelationship of round-spore-forming bacilli containing cell walls based on lysine and the non-spore-forming genera Caryophanon, Exiguobacterium, Kurthia, and Planococcus. Int J Syst Bacteriol. 1994; 44: 74-82. PubMed: https://pubmed.ncbi.nlm.nih.gov/8123563/
- Logan NA, Berkeley RCW. Identification of Bacillus strains using the API system. J Gen Microbiol. 1984; 130: 1871-1882. PubMed: https://pubmed.ncbi.nlm.nih.gov/6432953/
- El-Bondkly AM, El-Gendy MMAA, Wiese J, Imhoff JF. Phylogenetic diversity and antimicrobial activities of culturable endophytic actinobacteria isolated from different Egyptian marine sponges and soft corals. Australian Journal of Basic and Applied Sciences. 2012a; 6(4): 25-33.
- El-Bondkly AMA, El-Gendy MMAA, Bassyouni RH. Overproduction and biological activity of prodigiosin-like pigments from recombinant fusant of endophytic marine Streptomyces species. Antonie Van Leeuwenhoek. 2012b; 102: 719-734. PubMed: https://pubmed.ncbi.nlm.nih.gov/22777253/
- El-Gendy MMAA, El-Bondkly AMA. Evaluation and enhancement of heavy metals bioremediation in aqueous solutions by Nocardiopsis sp. MORSY1948, and Nocardia sp. MORSY2014. Brazilian Journal of Microbiology. 2016; 47: 571-586.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987; 4(4): 406-425.
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004; 101: 11030-11035. PubMed: https://pubmed.ncbi.nlm.nih.gov/15258291/
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018; 35: 1547-1549. PubMed: https://pubmed.ncbi.nlm.nih.gov/29722887/
- Cappuccino JG, Sherman N. Microbiology: A Laboratory manual. 4th Ed., Addison Wesley Longman, Inc. Harlow, England 1999; 199-204.
- Lavermicocca P, Valerio F, Visconti A. Antifungal activity of phenyl lactic acid against molds isolated from bakery products. Appl Environ Microbiol. 2003; 69: 634-640. PubMed: https://pubmed.ncbi.nlm.nih.gov/12514051/
- Mosmann T. Rapid colorimetric assays for cellular growth and survival: Application to proliferation and cytotoxicity assay. J Immunol Methods. 1983; 65: 55-63. PubMed: https://pubmed.ncbi.nlm.nih.gov/6606682/
- Wilson AP. Cytotoxicity and viability assays in animal cell culture: A Practical Approach. 3rd ed. (ed. Masters JRW) 2000; Oxford University Press.
- Ahmad J, Kakru D, Bali N, Lone S, Bashir H, Fomda B. Prevalence and risk factors for vancomycin resistant Enterococci isolated from clinical samples in Kashmir, North India: A hospital based study. J Adv Med Res. 2015; 12: 01-07.
- Tripathi A, Shukla SK, Singh A, Prasad KN. Prevalence, outcome and risk factor associated with vancomycin-resistant Enterococcus faecalis and Enterococcus faecium at a tertiary care hospital in Northern India. Indian J Med Microbiol. 2016; 34: 38-45. PubMed: https://pubmed.ncbi.nlm.nih.gov/26776117/
- Yadegarynia D, Roodsari SR, Arab-Mazar Z. Evaluation of antimicrobial susceptibility among Enterococcus species by E-test method at Khatamol Anbia hospital during 2013-2014. Archives of Clinical Infectious Diseases. 2016; 11: e60124.
- El-Gendy MMAA, El-Bondkly AMA, Keera AA, Ali AM. Incidence of methicillin-resistant Staphylococcus aureus (MRSA) in microbial community of cancer patients and evaluation of their resistant pattern. Arabian J Sci Engineering. 2018b; 43: 83-92.
- Sood S, Malhotra M, Das BK, Kapil A. Enterococcalinfections & antimicrobial resistance. Indian J Med Res. 2008; 128: 111-121. PubMed: https://pubmed.ncbi.nlm.nih.gov/19001673/
- Valentina Y, Umadevi S, Pramodhini S. Comparison of different phenotypic methods for detection of vancomycin resistant Enterococci (VRE) among clinical isolates. IP Int J Med Microbiol Trop Dis. 2020; 6: 123-125.
- Varghese V, Menon AR, Nair KP. Speciation and susceptibility pattern of Enterococcalspecies with special reference to high level gentamicin and vancomycin. J Clinic Diagn Res. 2020; 14: DC08- DC12.
- Freitas-Silva J, Silva-Oliveira T, Muricy G, Laport MS. Bacillus strains associated to Homoscleromorpha sponges are highly active against multidrug resistant bacteria. Curr Microbiol. 2020; 77: 807-815. PubMed: https://pubmed.ncbi.nlm.nih.gov/31925513/