More Information
Submitted: March 01, 2023 | Approved: March 14, 2023 | Published: March 15, 2023
How to cite this article: Trabelsi R, Yengui M, Mhaya A, Rebai A, Arpin C, et al. Detection of extended-spectrum betalactamase and carbapenemase-producing Enterobacteriaceae in Tunisia. Arch Biotechnol Biomed. 2023; 7: 001-011.
DOI: 10.29328/journal.abb.1001034
Copyright License: © 2023 Trabelsi R, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: UTIs; ESBL-producing; E. coli; K. pneumonia; CTX-M-type; Beta-lactamase and carbapenemase genes
Detection of extended-spectrum betalactamase and carbapenemase-producing Enterobacteriaceae in Tunisia
Rahma Trabelsi1* , Mariem Yengui1, Amel Mhaya2, Ahmed Rebai3, Corinne Arpin2 and Radhouane Gdoura1
, Mariem Yengui1, Amel Mhaya2, Ahmed Rebai3, Corinne Arpin2 and Radhouane Gdoura1
1Department of Life Sciences, Research Laboratory of Environmental Toxicology-Microbiology and Health (LR17ES06), Faculty of Sciences of Sfax, University of Sfax, BP 1171, 3000 Sfax, Tunisia
2University of Bordeaux, CNRS, MFP, UMR 5234, 146 rue Léo Saignat, Bordeaux Cedex, 33076, France
3Molecular and Cellular Screening Process Laboratory, Centre of Biotechnology of Sfax, Sfax, Tunisia
*Address for Correspondence: Dr. Rahma Trabelsi. Department of Life Sciences, Research Laboratory of Environmental Toxicology-Microbiology and Health (LR17ES06), Faculty of Sciences, University of Sfax, Tunisia, Email: [email protected]; [email protected]
The emergence of dramatic urinary tract infections (UTIs) caused by the members of the Enterobacteriales is an important public health problem in the community as well as in Tunisian hospitals. This study aims to investigate the prevalence of extended-spectrum β-lactamase (ESBL) and carbapenemase-producing uropathogenic isolates of Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae).
Based on decreased susceptibility to β-lactams antibiotics and analyzed for the presence of ESBL and carbapenemase genes by Real Time- polymerase chain reaction (RT-PCR), 56 uropathogenic isolates of E. coli (n = 36) and K. pneumoniae (n = 20) were confirmed positive for ESBLs. The CTX-M-type β-lactamases were mostly detected in E. coli isolates (21 strains, 58.33% [95% CI 38.09% - 72.06%]) followed by blaSHV-like (18 strains, 50% [95% CI 32.92% - 67.07%]), blaTEM-like and blaCMY--2-like simultaneously (15 strains, 41.67% [95% CI 25.51% - 59.24%]). Furthermore, the RT-PCR system on the K. pneumoniae strains demonstrated that blaSHV-12-like was the most predominant (16 strains, 80% [95% CI 56.33% - 94.26%]) followed by blaTEM-like (14 strains, 70% [95% CI 45.72% - 88.10%]), blaCTX-M belonging to groups 9 and 1 (11 strains, 55% [95% CI 31.52% - 76.94%]) and finally blaCMY--2-like (10 strains, 50% [95% CI 27.19% - 72.80%]). In addition, E. coli and K. pneumoniae strains harbored a carbapenemase gene blaOXA-48-like with 22.2% [95% CI 10.11% - 39.15%]; 20% [95% CI 12.83% - 43.66%], respectively.
Our results confirm the need to monitor the resistance to extended-spectrum β-lactams and to carbapenems among enterobacteria in Tunisia.
Urinary tract infections (UTIs) are a real public health problem both in terms of frequency and difficulty of treatment of multidrug-resistant bacteria, as well as of beta-lactamase and carbapenemase-producing bacteria [1] Extended-spectrum β-lactamase (ESBL) and Carbapenemase-producing Enterobacteriaceae (CPE) strains have been increasingly reported in Europe, South America, Asia, Oceania, and Africa in recent years [2]. However, E. coli and K. pneumoniae are also two of the most common pathogenic bacteria causing urinary tract and bloodstream infections by means of a remarkable range of virulence factors that can affect a wide variety of host cell processes [3]. The indiscriminate use of antibiotics has contributed to their significant presence in the environment, promoting resistant microorganisms by selection, mutation, and recombination through horizontal transfers [4]. The massive use of C3G antibiotics in the treatment of bacterial infections has contributed to the worldwide spread of Enterobacteriales, which produce ESBL [5]. In Tunisia, resistance to third-generation cephalosporins (C3G) has increased lately. In fact, of the total isolates of uropathogenic Enterobacteriales, it has been reported that 6.2% were resistant to C3G in 2012 and 19.7% in 2017 (https://www.infectiologie.org.tn/resistance.php). Until the late 1990s, the detected ESBLs were derived from the old narrow-spectrum penicillinases TEM-1, TEM-2, and SHV-1. Subsequently, other types of ESBLs have emerged as CTX-M, PER, VEB, and GES [6,7]. The CTX-M enzymes confer a high level of resistance to cefotaxime, although some variants, such as CTX-M-15, have also strong activity against ceftazidime [8]. Nowadays, Enterobacteriales may also express ESBLs that are not closely connected to TEM- or SHV-related species, including CTX-M and OXA-type ESBLs[9]. Currently, carbapenems are the most powerful agents prescribed for the treatment of serious infections caused by Enterobacteriales species given their broad spectrum of antimicrobial activity and excellent resistance to hydrolysis via most of the extended-spectrum b-lactamases (ESBLs) and cephalosporinases [10].
The main mechanism behind carbapenem resistance is the acquisition of OXA-48-like carbapenemases is an oxacillinase that is from the clinical isolate of K. pneumoniae in Turkey in 2001 [9,11]. These ESBLs are typically plasmid-mediated rather than chromosomally mediated β-lactamases [12]. Indeed, they have broader-range activity, covering carbapenems as well as extended-spectrum cephalosporins [13,14]. The resistance to broad-spectrum cephalosporins has increased among Enterobacterial strains from both human and animal sources [15]. Although Hall, et al. indicate that food might be a source of human-acquired antimicrobial-resistant E. coli due to the fact that similar ESBLs and plasmids encoding them have been detected in food-producing animals, food of animal origin, and humans [16]. In response to the emergence of ESBLs and carbapenemases in Tunisia [17], in addition to the misuse of antimicrobial agents, the launch of empirical treatment of UTIs cases is often based on the antimicrobial resistance pattern of the urinary pathogens from existing surveillance report [18]. Therefore, this study aims at developing an RT-PCR system for the detection of clinic blaTEM, blaSHV, blaCTX-M group-1, blaCTX-M group-9, blaCMY-2 and blaOXA-48 group genes in order to apply it in clinical E. coli and K. pneumoniae isolates collected from Tunisian hospitals. Herein, we describe the spread of E. coli and K. pneumoniae-harbored blaOXA-48 and ESBL-encoding genes in Tunisian hospitals.
Collection and identification of Enterobacteriales strains
Urine samples were collected in sterile containers using aseptic techniques from patients aged between 25 and 55 years and suspected to have UTIs. Of the total 1600 urine samples collected between January 2018 and March 2018, only 200 positive urine cultures were selected through random sampling. The selected urine samples were taken from patients of healthcare facilities and from community patients located in the city of Sfax and Tunis.
ESBL-producing Enterobacteriaceae (ESBL-E) identifi-cation was accomplished via conventional methods, including biochemical tests (BioMerieux API 10S) [19] and confirmed using Matrix-Assisted Laser Desorption Ionization-Time-Of-Flight/Mass Spectrometry (MALDI-TOF/MS, Brüker Daltonics).
Antibiotic susceptibility testing
All 176 Enterobacterial isolates were tested for antimicrobial susceptibility using the Mueller-Hinton (MH) agar (Bio-Rad) disk-diffusion technique. The susceptibility was tested to the following antibiotics: Amoxicillin-clavulanic acid (AMC), Ticarcillin (TIC), Cefotaxime (CTX), Ceftazidime (CAZ) Cefalotin (CF), Cefixime (CFM), Imipenem (IMP), Amikacin (AN), Gentamicin (GM), Netilmicin (NET), Tobramycin (NN), Nalidixic acid (NA), Ofloxacin (OFX), Ciprofloxacin (CIP), Fosfomycin (FFL), Nitrofurantoin (FM), Trimethoprim-sulfamethoxazole (SXT), Chloramphenicol (C). Inhibition zone diameters were interpreted according to Clinical and Laboratory Standards Institute recommendations and E. coli ATCC 25922 was used as a quality control strain [20].
Double disc synergy test
The isolates collected were distributed on an MH agar plate. The two antibiotic disks of CTX (30 µg) and CAZ (30 µg) were placed at a distance of 25 mm (edge to edge) from the AMC (20/10 µg) disc that was placed in the middle of the plate [21].
After a 24-h incubation, a test was considered positive if an enhanced zone of inhibition between either of the cephalosporin antibiotics and the AMC disc occurred. This indicated synergistic activity with Clavulanic acid and the presence of an ESBL [22].
Detection of extended-spectrum β-lactamase genes using Real-time PCR
The presence of different Ambler class A β-lactamase encoding genes has been tested through RT-PCR using specific primers for blaCTX-M, blaSHV, and blaTEM [19]. Other β-lactamase encoding genes, belonging to class D (blaOXA-48-like) and class C (blaCMY--2-like), have also been tested as previously described [23,24].
RT-PCR was performed for beta-lactamase genes of the family TEM-like, SHV-like, and CTX-M of the two most prevalent groups, ie. CTX-M of group 1 and group 9 (named CTX-M-G1 and CTX-M-G9, respectively), CMY-2-like, and OXA-48-like via specific primers, designed using Primer-BLAST (Table 1) Figure 1.
| Table 1: List of primers used for real-time PCR amplification of ESBLs and carbapenemase genes. | ||||
| Genes | Primer sequence (5'–›3') | T°m | Product size(bp) | GenBank Accession no. |
| SHV-like | FW: AGCCGCTTGAGCAAATTAAA | 59.99 | 77 | LC229232.1 |
| RV: GCTGGCCAGATCCATTTCTA | 60.18 | |||
| TEM-like | FW: GATAAATCTGGAGCCGGTGA | 60.04 | 78 | MG860488.1 |
| RV: GATACGGGAGGGCTTACCAT | 60.17 | |||
| CTX-M- G1(blaCTX-M15-like) | FW: CACCAATGATATTGCGGTGA | 60.34 | 77 | MG288677.1 |
| RV: GTTGCGGCTGGGTAAAATAG | 59.61 | |||
| CTX-M- G9(blaCTX-M9-like) | FW: TACTTCACCCAGCCTCAACC | 60.11 | 78 | CP028990.1 |
| RV: ACCGTCGGTGACGATTTTAG | 59.99 | |||
| CMY-2-like | FW: CCAGAACTGACAGGCAAACA | 59.87 | 65 | LC229227 |
| RV: CCTGCCGTATAGGTGGCTAA | 60.11 | |||
| OXA-48-like | FW: GTAGTCAGCGCATCGTGAAA | 60.02 | 73 | MN654469.1 |
| RV: CCCGTTTTAGCCCGAATAAT | 60.12 | |||
| RV: Reverse; FW: Forword; T°m: Melting temperature; PCR: Polymerase Chain Reaction; ESBLs: Extended-Spectrum β-lactamases | ||||
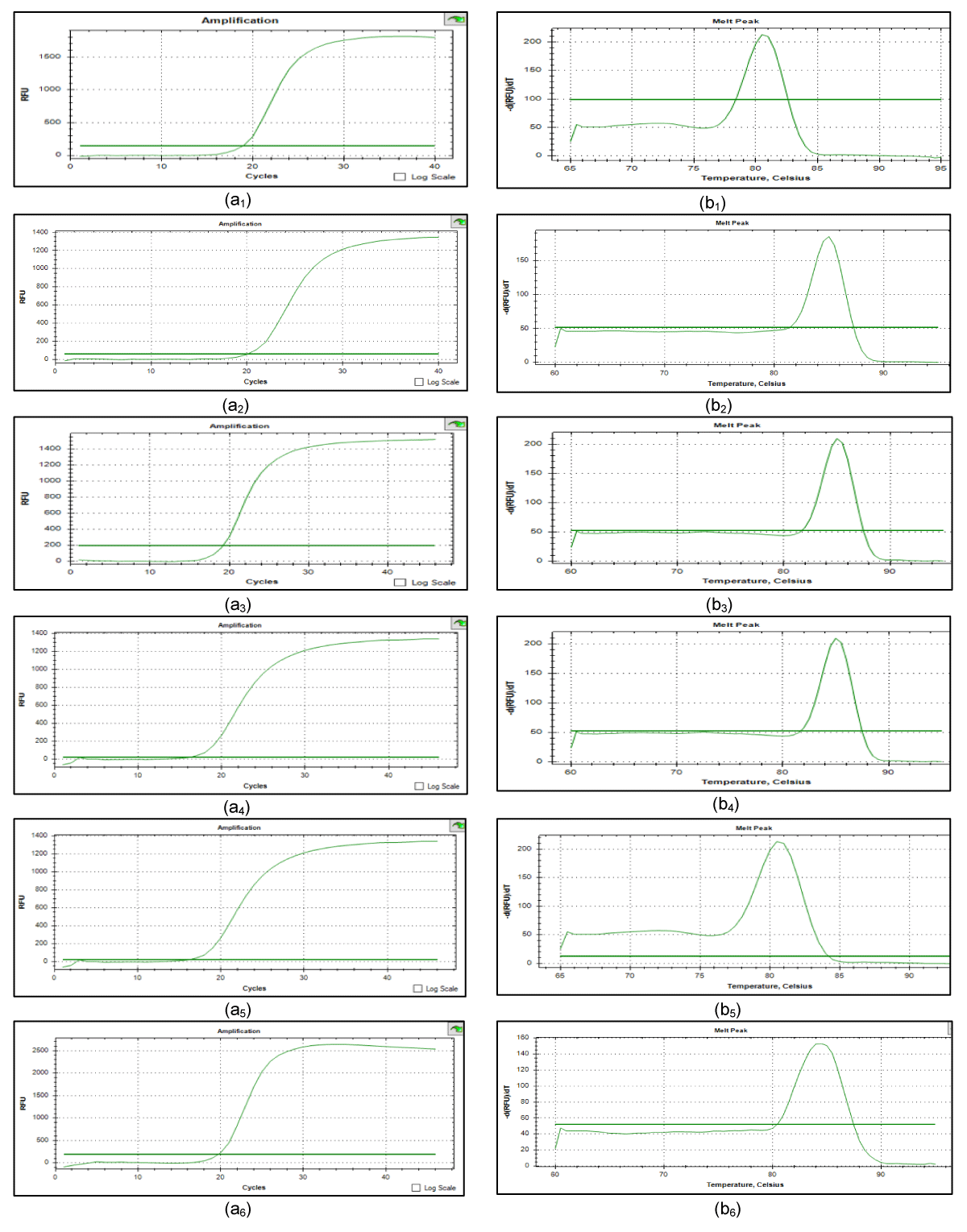
Figure 1: Results of real time PCR for blaSHV-12-like/ blaTEM-like/ blaCTX-M-G1/ blaCTX- M-G9/ blaCMY--2-like / blaOXA-48-like (a1), (a2), (a3), (a4), (a5), (a6): the profile of DNA amplification for blaSHV-12-like, blaTEM-like, blaCTX-M-G1, blaCTX-M-G9, blaCMY--2-like, blaOXA-48-like respectively. (b1), (b2), (b3), (b4), (b5), (b6): the profile of melt curve for blaSHV-12-like (81), blaTEM-like (85.5), blaCTX-M-G1 (83), blaCTX-M-G9 (85), blaCMY--2-like (80.5), blaOXA-48-like (84.5) respectively.
Monoplex PCR assays were optimized using Applied Biosystems including the Taq SYBR Green MasterMix (Invitrogen Life Technologies) and Microamp 96-Well Reaction Plate 0.1mL (Thermo-Fisher Scientific). Briefly, reactions were performed in a final volume of 20μL containing 100nM DNA solution and 10 µmol of each gene-specific primer. PCR amplification conditions were as follows: initial denaturation step at 95 °C for 3 min; 45 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, extension at 72 °C for 2 min, followed by a final extension step at 72 °C for 10 min. Amplification results were analyzed with the threshold and baseline set between 10 and 25 cycles. Melting-curve analysis was performed as follows: 60 °C for 3s (ramp rate = 5 °C/s) until reaching a final temperature of 95 °C, with fluorescence fluctuation, analyzed during the latter. The number of DNA molecules present in the sample was determined based on the amount of fluorescence detected by the RT-PCR instrument [23]. This quantified fluorescence resulted in a Cycle Threshold value (Ct) that corresponds to the number of amplification cycles required to obtain a given amount of DNA [25].
Statistical analysis
Statistical tests including Spearman's rank correlation analysis, χ2 test, and multiple logistic regression analysis were used to evaluate the associations between the determinant bla group genes and the various levels of antibiotic resistance phenotypes. The level of significance was set at p < 0.05 in this study. All statistical analyses were carried out using IBM SPSS Statistics for Windows, Version 16.0 (SPSS Inc., Chicago, IL, USA).
Prevalence of ESBL-producing Enterobacteriaceae in Tunisian healthcare facilities
Of the 176 isolates that were classified as Enterobacteriales, 100 isolates (50%) were identified to be E. coli, which was predominant, followed by 50 isolates of K.pneumoniae (25%), 20 isolates of Enterobacter cloacae complex (10%) and 6 isolates of Citrobacter koseri (3%). Fifty-six isolates (28%), considered ESBL-producing, were initially classified as Enterobacter spp. through biochemical tests (API 10S gallery). Following matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS), those isolates were re-identified and confirmed as 36 strains of E. coli and 20 of K. pneumoniae.
Antibiotic resistance patterns
The antimicrobial susceptibility patterns were determined for all 176 Enterobacteriales urine isolates. After identification, the total of 56 ESBL-producing uropathogenic isolates included 36 strains of E. coli and 20 of K. pneumoniae.
The ESBL-producing E. coli and K. pneumoniae uropathogenic isolates showed higher levels of resistance to all antibiotics compared to the non-ESBL-producing isolates except for imipenem, since the majority of the isolates tested in this study were imipenem-sensitive. Antibiotic susceptibility testing for the 56 isolates is summarized in Table 2.
A high prevalence of resistance was observed against third-generation antibiotics cephalosporins (3GCs). All the isolates were resistant to AMX, TIC, CAZ, and CTX. Resistance to 3GCs was associated predominantly with the presence of blaCTX-M genes.
The 36 isolates of E. coli showed a high level of resistance to CIP (n = 34, 94.44% [CI 81.33% - 99.31%]), OFX (n = 33, 91.66% [CI 77.53% - 98.24%]), as well as a moderate level of resistance to imipenem (n = 7, 19.45% [CI 8.1% - 36%]) (Table 2). The 20 isolates of K. pneumoniae also showed a high level of resistance to members of the quinolone family, including CIP (n = 16, 80% [CI 56.33% - 94.26%]), OFX (n = 15, 75% [CI 50.89% - 91.34%]), in addition to imipenem which was reported on a level of four K. pneumoniae strains (20% [CI 5.73% - 43.66%]) (Table 2). All isolates were susceptible to colistin. The double-disk synergy test showed that 29% of the isolates were ESBL-positive.
All ESBL-producing E. coli strains showed a decreased susceptibility to ß-lactams, except for cephamycins (cefoxitin) and carbapenems. This latter remained susceptible to fosfomycin, and both were resistant to nitrofurantoin. All E. coli isolates were resistant to co-trimoxazole (sulfamethoxazole-trimethoprim). They exhibited different aminoglycoside phenotypes (resistance to gentamycin, tobramycin, and amikacin, 26%, 47%, and 20%, respectively) (Table 2). Among the K. pneumoniae strains, various resistance profiles were demonstrated to tested antibiotics (Table 2).
| Table 2: Antibiotic susceptibility profile of ESBL-producing E. coli and K. pneumonia. | ||||||
| Beta-lactams | Antibiotics | ESBL-producing E.coli | ESBL-producing K. pneumoniae | |||
| Resistors(n = 36); [95% CI] | Sensitive; [95% CI] | Resistors(n = 20); [95% CI] | Sensitive; [95% CI] | |||
| Penicillin | AMC | 100% (36) | 0 | 100% (20) | 0 | |
| TIC | 100% (36) | 0 | 100% (20) | 0 | ||
| Cephalosporin (1GCs) | CF | 80.55% (29) [63.97%-91.80%] |
19.45% (7) [8.1%-36%] |
100% (20) | 0 | |
| Cephalosporin (3GCs) | CFM | 72.22% (26) [54.81%-85.79%] |
27.78% (10) [14.20%-45.18%] |
90% (19) [75.12%-99.87%] |
10% (1) [0.12%-24%] |
|
| CTX | 100% (36) | 0 | 100% (20) | 0 | ||
| CAZ | 100% (36) | 0 | 100% (20) | 0 | ||
| Carbapenem | IMP | 19.45% (7) [8.1%-36%] |
80,55% (29) [63.97%-91.80%] |
20% (4) [5.73%-43.66%] |
80% (16) [56.33%-94.26%] |
|
| Fluoroquinolone | OFX | 91.66% (33) [77.53%-98.24%] |
8.34% (3) [77.53%-98.24%] |
75% (15) [50.89%-91.34%] |
25% (5) [8.65%-49.10] |
|
| CIP | 94.44% (34) [81.33%-99.31%] |
5.56% (2) [0.68%-18.66%] |
80% (16) [56.33%-94.26%] |
20% (4) [5.73%-43.66%] |
||
| Aminosides | AN | 27.78% (10) [14.20%-45.18%] |
72.22% (26) [54.81%-85.79%] |
20% (4) [5.73%-43.66%] |
80% (16) [56.33%-94.26%] |
|
| GM | 47.22% (17) [30.40%-64.51] |
52.78% (19) [35.48%-69.59%] |
70% (14) [45.72%-88.10%] |
30% (6) [75.12%-99.87%] |
||
| NET | 30.55% (11) [16.34%-48.10%] |
69.45% (25) [54.81%-85.79%] |
45% (9) [23.05%-68.47%] |
55% (11) [31.52%-76.94%] |
||
| NN | 47.22% (17) [30.40%-64.51%] |
52.78% (19) [35.48%-69.59%] |
45% (9) [23.05%-68.47%] |
55% (11) [31.52%-76.94%] |
||
| FFL | 5.56% (2) [0.68%-18.66%] |
94.44% (34) [81.33%-99.31%] |
40% (8) [19.11%-63.94%] |
60% (12) [36.05%-80.88%] |
||
| SXT | 36.11% (13) [20.82%-53.77%] |
63.86% (23) [46.22%-79.17%] |
65% (13) [40.78%-84.60%] |
35% (7) [15.39%-59.21%] |
||
| FM | 13.88% (5) [4.66%-29.49%] |
86.12% (31) [70.50%-95.33%] |
35% (7) [15.39%-59.21%] |
65% (13) [40.78%-84.60%] |
||
| CI: Confidence Intervals; 1GCs: First-Generation; 3GCs: Third-Generation. AMC: Amoxicillin - clavulanic acid; TIC: Ticarcillin; CTX: Cefotaxime; CAZ: Ceftazidime; CF: Cefalotin; CFM: Cefixime; IMP: Imipenem; AN: Amikacin; GM: Gentamicin; NET: Netilmicin; NN: Tobramycin; OFX: Ofloxacin; CIP: Ciprofloxacin; FFL: Focfomycin; FM: Nitrofurantoin; SXT: Trimethoprim-sulfamethoxazole | ||||||
Prevalence of ESBL genes
The newly developed RT-PCR system was efficient in detecting the bla group genes. All positive phenotypic ESBL and carbapenemase isolates were analyzed for the presence of genes encoding blaTEM-like, blaSHV-like, blaCTX-M-G1, blaCTX-M-G9, blaCMY--2-like and blaOXA-48-like. Further details on the results are shown in Tables 3,4.
| Table 3: Prevalence of blaTEM-like, blaSHV-like, blaCTX-M1, blaCTX-M9, blaCMY--2-like and blaOXA-48-like genes in ESBL producers for E.coli and K. pneumoniae. | ||
| Identified genes | E.coli [95% CI] | K.pneumoniae [95% CI] |
| SHV-like | 50%[32.92%-67.07%] | 80%[56.33%-94.26%] |
| TEM-like | 41.67%[25.51%-59.24%] | 70%[45.72%-88.10%] |
| CTX-M-G9 | 58.33%[38.09%-72.06%] | 55%[31.52%-76.94%] |
| CTX-M-G1 | 58.33%[38.09%-72.06%] | 55%[31.52%-76.94%] |
| CMY-2-like | 41.67%[25.51%-59.24%] | 50%[27.19%-72.80%] |
| OXA-48-like | 22.2%[10.11%-39.15%] | 20%[12.83%-43.66%] |
| SHV-like/TEM-like | 30.56%[16.34%-48.10%] | 50%[27.19%-72.80%] |
| SHV-like/TEM-like/CTX-M-G9/CTX-M-G1 | 16.67%[6.37%-32.81%] | 40%[19.11%-63.94%] |
| CTX-M-G9/CTX-M-G1/OXA-48-like | 8.33%[27.19%-72.80%] | 10%[1.23%-31.69%] |
| CTX-M-G9/CTX-M-G1 | 41.67%[27.19%-72.80%] | 45%[23.05%-68.47%] |
| OXA-48-like/CMY-2-like | 13.89%[4.66%-29.49%] | 15%[3.20%-37.89%] |
| CTX-M-G9/CMY-2-like | 19.44%[8.19%-36.02%] | 25%[8.65%-49.10%] |
| CTX-M-G1/CMY-2-like | 22.22%[10.11%-39.15%] | 25%[8.65%-49.10%] |
| CTX-M-G9/OXA-48-like | 11.11%[3.11%-26.06%] | 10%[1.23%-31.69%] |
| CTX-M-G1/OXA-48-like | 11.11%[3.11%-26.06%] | 10%[1.23%-31.69%] |
| CMY-2-like/SHV-12-like | 16.67%[6.37%-32.81%] | 40%[19.11%-63.94%] |
| CI: Confidence Intervals | ||
| Table 4: Distribution of bla genotypes among the clinical K. pneumoniae and E.coli strains and their respective resistance rates to CTX, CAZ, CF, CFM and IMP. | ||||||||||||||
| E.coli | K.pneumoniae | |||||||||||||
| % Resistance to | % Resistance to | |||||||||||||
| Genotype | Number of isolates (n=36) | Percentage (%) |
CTX | CAZ | CF | CFM | IMP | Number of isolates (n=20) | Percentage (%) |
CTX | CAZ | CF | CFM | IMP |
| SHV-like | 18 | 50 | 100 | 100 | 66.66 | 66.66 | 27.7 | 16 | 80 | 100 | 100 | 100 | 87.5 | 25 |
| TEM-like | 15 | 41.67 | 100 | 100 | 73.3 | 53.33 | 13.33 | 14 | 70 | 100 | 100 | 100 | 85.7 | 21.4 |
| CTX-M-G9 | 21 | 58.33 | 100 | 100 | 90.4 | 76.19 | 14.28 | 11 | 55 | 100 | 100 | 100 | 100 | 27.27 |
| CTX-M-G1 | 21 | 58.33 | 100 | 100 | 85.57 | 76.19 | 9.52 | 11 | 55 | 100 | 100 | 100 | 100 | 27.27 |
| CMY-2-like | 15 | 41.67 | 100 | 100 | 86.66 | 73 | 26.66 | 10 | 50 | 100 | 100 | 100 | 100 | 30 |
| OXA-48-like | 8 | 22.2 | 100 | 100 | 87.5 | 62.5 | 100 | 4 | 20 | 100 | 100 | 100 | 90 | 100 |
| SHV-like/TEM-like | 11 | 30.56 | 100 | 100 | 72.7 | 54.45 | 18.18 | 10 | 50 | 100 | 100 | 100 | 100 | 30 |
| SHV-like/TEM-like/CTX-M-G9 /CTX-M-G1 | 6 | 16.77 | 100 | 100 | 100 | 100 | 33.33 | 8 | 40 | 100 | 100 | 100 | 100 | 37.5 |
| CTX-M-G9/CTX-M-G1/OXA-48-like | 3 | 8.33 | 100 | 100 | 66.66 | 33.33 | 66.66 | 2 | 10 | 100 | 100 | 100 | 100 | 100 |
| CTX-M-G9/CTX-M-G1 | 15 | 41.67 | 100 | 100 | 100 | 93.3 | 11.33 | 9 | 45 | 100 | 100 | 100 | 100 | 33.33 |
| OXA-48-like/CMY-2-like | 5 | 13.89 | 100 | 100 | 80 | 80 | 80 | 3 | 15 | 100 | 100 | 100 | 100 | 100 |
| CTX-M-G9/CMY-2-like | 7 | 19.44 | 100 | 100 | 100 | 85.7 | 14.28 | 5 | 25 | 100 | 100 | 100 | 100 | 60 |
| CTX-M-G1/CMY-2-like | 8 | 22.2 | 100 | 100 | 87.5 | 75 | 14.28 | 5 | 25 | 100 | 100 | 100 | 100 | 60 |
| CTX-M-G9/OXA-48-like | 4 | 11.11 | 100 | 100 | 75 | 25 | 75 | 2 | 10 | 100 | 100 | 100 | 100 | 100 |
| CTX-M-G1/OXA-48-like | 4 | 11.11 | 100 | 100 | 75 | 50 | 75 | 2 | 10 | 100 | 100 | 100 | 100 | 100 |
| CMY-2-like/SHV-like | 6 | 16.67 | 100 | 100 | 66.66 | 66.66 | 50 | 8 | 40 | 100 | 100 | 100 | 100 | 25 |
| CTX: Cefotaxime; CAZ: Ceftazidime; CF: Cefalotin; CFM: Cefixime; IMP: Imipenem | ||||||||||||||
The ESBL production was detected in all ceftazidime and cefotaxime-resistant E. coli and K. pneumoniae strains recovered in this study. In terms of E. coli, blaCTX-M-G1, and blaCTX-M-G9 were the most prevalent (21 strains, 58.33% [95% CI 38.09% - 72.06%]) followed by blaSHV-like (18 strains, 50% [95% CI 32.92% - 67.07%]), blaTEM-like and blaCMY--2-like simultaneously (15 strains, 41.67% [95% CI 25.51% - 59.24%]). Furthermore, the RT-PCR system on the K. pneumoniae strains demonstrated that blaSHV-like was the most predominant (16 strains, 80%[95%CI 56.33% - 94.26%]) followed by blaTEM-like (14 strains, 70% [95% CI 45.72% - 88.10%]), blaCTX-M-G9 and blaCTX-M-G1 (11 strains, 55% [95% CI 31.52% - 76.94%]) and finally blaCMY--2-like (10 strains, 50% [95% CI 27.19% - 72.80%]) (Tables 3,4).
On the other hand, 22.2% (8 strains) and 20% (4 strains) of ESBL-producing E. coli and K. pneumoniae strains, respectively, possessed at least one bla group gene with 6 different genotypes (Tables 3, 4).
Besides all the third-generation cephalosporin-resistant strains, 14 of them (6 E. coli, 16.64% and 8 K. pneumoniae, 40%, respectively,) harbored the four bla groups genes: ie. blaTEM-like; blaSHV-like, and both blaCTX-MG1 and blaCTX-M-G9.
Interestingly, the presence of the blaOXA-48-like gene was reported in eight isolates of E. coli (22.2% [95% CI 10.11% - 39.15%] and four isolates of K. pneumoniae (20% [95% CI 12.83% - 43.66%]) in this study. Indeed, seven E. coli and four K. pneumoniae isolates, resistant to imipenem, were found to produce OXA-48-like (Table 4). These data confirmed that the imipenem-resistant K. pneumoniae carrying the blaOXA-48 gene was widespread in Tunisian healthcare facilities.
In conclusion, a high CTX-M prevalence was found in ESBL-producing strains of both E. coli and K. pneumonia. Nevertheless, all the OXA-48-like-producing isolates coproduced other β-lactamases such as TEM-like, SHV-like, CTX-M, and CMY-2-like (Tables 3,4).
Resistance to three or more β-lactam agents was associated with specific gene types and numbers
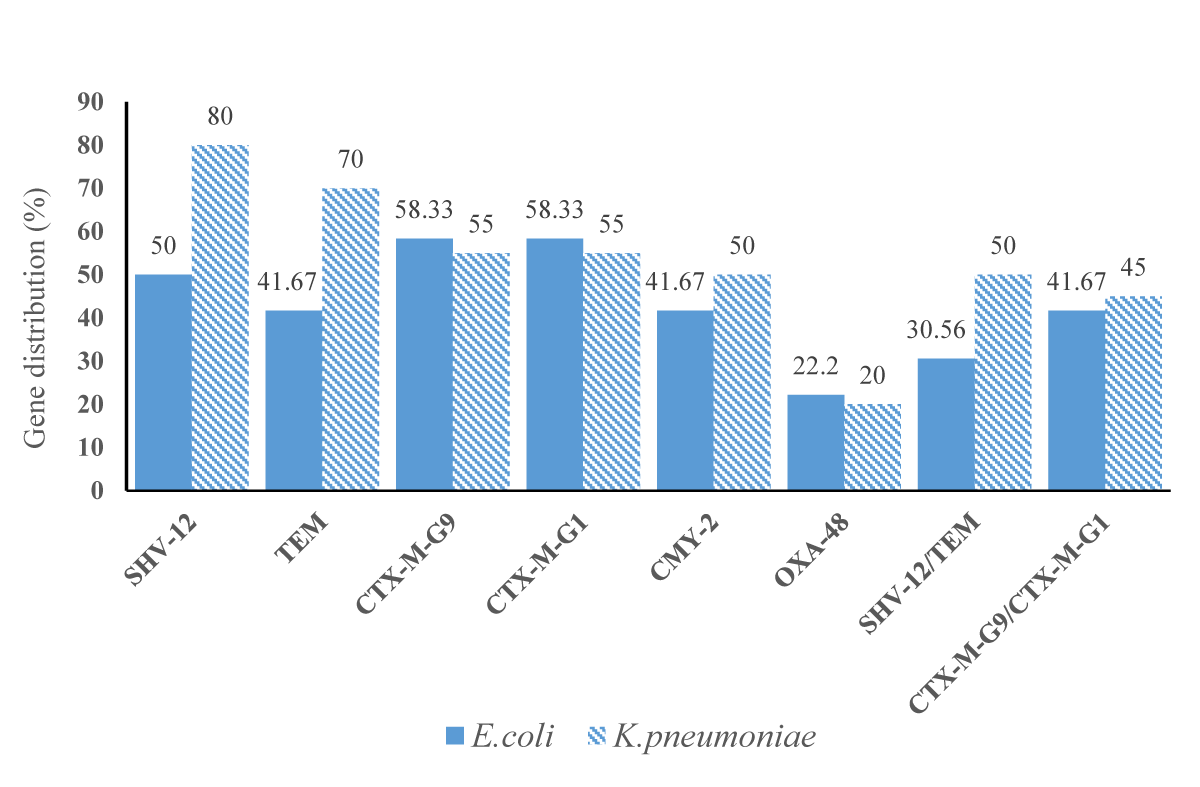
Resistance to three or more agents was associated with the presence of a specific gene type. Specifically, analysis of our data revealed that carriage of blaCMY--2-like and blaOXA-48-like genes correlated positively with resistance to three or more antibiotics (Po = 0.049 and 0.005, respectively; Table 5). Many strains harboring blaOXA-48 group genes also co-harbored blaCMY--2-like, blaCTX-M-G1, and/or blaCTX-M-G9 genes (Table 4 and Figure 2). This positive association between blaOXA-48-like and/or blaCMY--2-like gene(s) and the resistance to three or more antibiotics was further confirmed by multiple logistic regression analysis (Table 6).
Figure 2: Distribution of the six group genes in the 36 E. coli and 20 K. pneumoniae clinical strains.
| Table 5: Relationship between bla genes and resistance to three or more antibiotics in the 56 clinical strains (by χ2 test). | ||||||||||||||||||
| Resistance rate % (positive/total) | ||||||||||||||||||
| blaSHV-like | blaTEM-like | blaCTX-M9 | blaCTX-M15 | blaCMY--2-like | blaOXA-48-like | |||||||||||||
| Antibiotics | + | - | p -value | + | - | p -value | + | - | p -value | + | - | p -value | + | - | p -value | + | - | p -value |
| Resistance to >3 agents | 29/35 | 5/21 | 0.543 | 21/28 | 4/28 | 0.313 | 29/34 | 6/22 | 0.248 | 28/33 | 6/23 | 0.3 | 23/25 | 9/31 | 0.049 | 11/11 | 11/45 | 0.005 |
| Table 6: Relationship between bla genes and resistance to three or more antibiotics in the 56 clinical strains (by multiple logistic regression analysis). | ||||||||||||||||||
| Resistance rate % (positive/total) | ||||||||||||||||||
| blaTEM-like | blaSHV-like | blaCTX-M9 | blaCTX-M15 | blaCMY-2-like | blaOXA-48-like | |||||||||||||
| Antibiotics | X2 | p -value | OR (95% CI) |
X2 | p -value | OR (95% CI) |
X2 | p -value | OR (95%CI) |
X2 | p -value | OR (95% CI) |
X2 | p -value | OR (95 % CI) |
X2 | p -value | OR (95% CI) |
| Resistance to >3 agents | - | - | - | - | - | - | -- | - | - | - | - | - | 3.072 | 0.054 | 4.7 (0.913-24.250) |
- | - | - |
In this study, we characterized ESBL- and carbapenemase-producing E. coli and K. pneumoniae clinical isolates collected from Tunisian community and healthcare facilities patients suffering from UTIs. The presence of different Ambler class A β-lactamase encoding genes has been tested by means of RT-PCR with specific primers for blaCTX-M, blaSHV, and blaTEM [26]. Other β-lactamase encoding genes belonging to class D (blaOXA-48) and class C (blaCMY-2) have also been tested as previously described [23,27].
Some previous studies have also affirmed the emerging problem of ESBL-producing E. coli and K. pneumoniae isolates in different Mediterranean countries [28].
In this 3-month survey, the incidence rate of ESBLs-producing Enterobacteriales in urine samples taken from UTI patients was 28.72%. The antimicrobial susceptibility patterns were determined for all the E. coli and K. pneumoniae urine isolates collected. The ESBL-producing isolates showed higher levels of resistance to all antibiotics compared to the non-ESBL-producing isolates. Indeed, data showed that 20% of ESBL-producing E. coli and K. pneumoniae isolates were imipenem-resistant, due to their co-production with an OXA-48-like carbapenemase. This is unsurprising given that ESBL-producing Enterobacteriaceae strains are frequently associated with co-resistance to other antimicrobial agents such as aminoglycosides and fluoroquinolones [29]. The ESBL-producing isolates showed the highest levels of resistance to ampicillin, ceftazidime, and ciprofloxacin. These findings are in accordance with many previous reports [30]. Moreover, ESBLs producers have been proven to be resistant, not only to beta-lactam agents, but also to other antimicrobial agents such as tetracycline, fluoroquinolones, aminoglycosides, and trimethoprim/sulfamethoxazole [31]. Similar resistance profiles were also reported in the present study. There are so many factors responsible for such an elevated rate of antibiotic resistance, some of which are the massive and improper use of antimicrobial agents by health professionals in hospitals, in addition to the self-prescription by community patients [32].
E. coli and K. pneumoniae ESBL-producing isolates from UTIs are increasingly found worldwide, causing serious problems in many countries. Findings from the present study showed that 64% of E. coli and 35% of K. pneumoniae isolates were ESBL-producing bacteria. These findings are consistent with those of Bazaz, et al. who reported a prevalence of ESBLs in K. pneumoniae and E. coli bacteria as high as 59.2%. Reports on ESBL production rate in Enterobacteriales isolates from different countries also show significant variations [33]. In Japan and USA, the prevalence rate of ESBL production in Enterobacteriaceae was reported to be 40 and 44%, respectively [31].
In this study, CTX-M enzymes were the dominant type of ESBLs. Still, the data shows clearly that blaCTX-M-G1 was the most frequently detected in E. coli and K. pneumoniae isolates (58.33% and 55%, respectively). As well, the blaCTX-M<-G9 was detected with a 55% rate. This pattern of results confirms earlier findings showing that the predominance of CTX-M-15 and CTX-M-27-producing ESBLs from human Enterobacteriales isolates is widely present in Tunisia [34]. In addition, these results correlate fairly well with previous studies showing that blaCTX-M-15 and blaCTX-M 14 are the most common genes responsible for mediating extended-spectrum cephalosporin resistance in these isolates. Indeed, Mamlouk, et al. detected the blaCTX-M-15 enzyme in 30% of Enterobacteriales (35% from E. coli and 27% from K. pneumoniae) collected from different wards of Charles Nicolle Hospital in Tunis [35]. More recently in 2014, Ferjani, et al. reported that 88% of cefotaxime-resistant E. coli strains, isolated from the urine of patients in a Tunisian hospital, harbored the blaCTX-M-15 gene [36]. These studies confirm the current spread of the CTX-M-15 encoding gene, which encodes the most prevalent β-lactamase detected among ESBLs-positive K. pneumoniae and E. coli strains in Tunisian hospitals. The increased use of cefotaxime and ceftazidime might have contributed to the emergence of ESBLs, particularly these CTX-M-type enzymes. Several previous studies have also discussed the emerging problem of ESBL-producing E. coli and K. pneumoniae isolates in different geographic regions, including the Mediterranean basin [15] and North Africa [37]. Results of the present study confirm that these CTX-M-type enzymes are the most dominant ESBL type in clinical E. coli and K. pneumoniae isolates in Tunisia [38]. Interestingly, a similar pattern of results was found in other countries [39].
According to previous reports, the emergence of CTX-M-15 and CTX-M-27 ESBLs from human Enterobacteriales isolates has been increasingly reported in many countries around the world, including Tunisia [39].
Data are consistent with those described in the literature [40]. Few studies on molecular analysis of ESBLs-E from Urinary Tract infections were conducted in Tunisia [40]. Currently, CTX-M enzymes have replaced the traditional ESBL types, such as SHV and TEM enzymes, as the most prevalent ESBL type [8].
Hence, the present study is the first to report on the coexistence of CTX-M-G1 and CTX-M-G9 enzymes in Tunisian ESBL-positive E. coli and K. pneumoniae uropathogens isolates.
Similarly, in North Africa, many studies have confirmed the increase of ESBL-producing Enterobacteriales from urine [38]. In Morocco, Girlich, et al. reported a high rate of ESBL-producing Enterobacteriales at the university hospital [41]. Likewise, in Egypt, the CTX-M-15 encoding gene has been detected in clinical isolates of E. coli from Cairo [42].
The isolates of K. pneumoniae and E. coli are common causes of nosocomial infections such as UTIs. These latter can lead to renal failure, if left undiagnosed or late diagnosed. β-Lactam is one of the most effective drugs in UTI treatment. ESBL bacteria, with inactivation of a wide range of β-lactam drugs, especially cephalosporins, and monobactam, can cause treatment failure and increase healthcare costs. Therefore, the study of gene resistance to β-lactamase is important. It seems that the emergence and spread of these bacteria are due to prolonged hospitalization, increased consumption of β-lactam antibiotics (especially ceftazidime), use of catheters, and experimental treatments against antibiotic-resistant [43].
Interestingly, the present study reported the presence of the blaOXA-48-like gene in eight isolates of E. coli (22.2%) and four isolates of K. pneumoniae (20%). In addition, seven imipenem-resistant E. coli and four K. pneumoniae isolates were found to produce OXA-48-like. Several studies on the emergence of OXA-48-producing Enterobacteriales have been carried out in Tunisia.
For example, Ktari, et al. [44] reported the spread of 21 (13.7%) K. pneumoniae isolates producing the blaOXA-48-like encoding-gene in a Tunisian university hospital. In 2012, Saidani, et al. [17] examined 21 ESBLs-producing Enterobacteriaceae, with reduced susceptibilities to carbapenems, and found that 5 out of the 21 isolates investigated were OXA-48-like positive.
Indeed, a current study by Mhaya, et al. [45] reports the first description of K. pneumoniae carrying the carbapenemase NDM-1 and OXA-like-1 in the Tunisian community setting and confirms that the NDM-1/OXA-like -1-positive ST147 K. pneumoniae clone has become endemic in this country, the strain showed the presence of blaCTX-M-G1 and blaNDM genes together with blaTEM-like, blaSHV-like and blaOXA-1-like genes. This suggests that NDM-1/OXA-48-like-producing ST147 K. pneumoniae, although reported as an emerging clone in Tunisia.
More recently, Charfi, et al. [46] reported that among enterobacterial clinical isolates recovered in the Center of Maternity and Neonatology of Monastir in Tunisia, one tested positive for the OXA-48 gene that co-expressed the blaCTX-M-15 gene.
In this study, 5/56 OXA-48-like isolates additionally expressed the blaCTX-M- G1 and blaCTX-M-G2 genes. In 8/56 cases, we noted the presence of the combination of two β-lactamases (OXA-48-like and CMY-2-like), highlighting the common combination of several β-lactamases in a single isolate. Since oxacillinases produced alone possess weak activity against carbapenems, the co-existence of two or more β-lactamases has become a common bacterial strategy to increase antimicrobial resistance [46].
These findings are of great concern given the fact that the rapid propagation of multidrug-resistant bacteria, particularly cephalosporin and carbapenem-resistant strains, represents a major therapeutic and epidemiological threat. Therefore, the implementation of strict precautionary measures for infection prevention, in addition to regular surveillance studies, is urgently needed in order to limit the increasing spread of these multidrug-resistant bacteria.
In conclusion, this study described the high prevalence of CTX-M- and OXA-48-like-producing of E. coli and K. pneumoniae uropathogenic isolates in Tunisian healthcare facilities. However, our results, demonstrate the correlation between genetic and phenotypic profiles of ESBL-producing members of Enterobacteriales. The study highlights an escalating crisis of cephalosporins and carbapenemase resistance in Enterobacteriales, causing UTIs.
Author contributions statement
1. Rahma Trabelsi contributed to this investigation and carried out most of the experimental procedures. She actively participated in data acquisition, analysis, and interpretation and wrote the article.
2. Mariem Yengui contributed with her efforts and expertise.
3. Amel Mhaya has provided essential reagents and critical revision of the manuscript for important intellectual content.
4. Ahmed Rebai was responsible for the conception, design, and development of the experiments.
5. Corinne Arpin was responsible for the conception, design, and development of the experiments.
6. Radhouane Gdoura was responsible for the conception, design, and development of the experiments. All authors contributed to the drafting of the manuscript and gave their final approval to the submitted version.
We are grateful to Dr. Salem Riguane (Head of the private lab for medical analysis), Dr. Gazi Boujelben (Head of the lab for medical analysis at the Aliya Clinic, Sfax), and Pr. Dr. Olfa Bahri (Bacteriology laboratory at Hospital Aziza Othmena, Tunis) for their valuable contribution to the success of this study. This work was supported by the Tunisian Ministry for Higher Education and Scientific Research.
- Lee DS, Lee SJ, Choe HS. Community-Acquired Urinary Tract Infection by Escherichia coli in the Era of Antibiotic Resistance. Biomed Res Int. 2018 Sep 26;2018:7656752. doi: 10.1155/2018/7656752. PMID: 30356438; PMCID: PMC6178185.
- Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect. 2014 Sep;20(9):821-30. doi: 10.1111/1469-0691.12719. PMID: 24930781.
- Shakya P, Shrestha D, Maharjan E, Sharma VK, Paudyal R. ESBL Production Among E. coli and Klebsiella spp. Causing Urinary Tract Infection: A Hospital Based Study. Open Microbiol J. 2017 Apr 28;11:23-30. doi: 10.2174/1874285801711010023. PMID: 28553414; PMCID: PMC5427687.
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010 Sep;74(3):417-33. doi: 10.1128/MMBR.00016-10. PMID: 20805405; PMCID: PMC2937522.
- Poirel L, Pitout JD, Nordmann P. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2007 Oct;2(5):501-12. doi: 10.2217/17460913.2.5.501. PMID: 17927473.
- Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. 2014; 18:1–36.
- Jena J, Sahoo RK, Debata NK, Subudhi E. Prevalence of TEM, SHV, and CTX-M genes of extended-spectrum β-lactamase-producing Escherichia coli strains isolated from urinary tract infections in adults. 3 Biotech. 2017 Aug;7(4):244. doi: 10.1007/s13205-017-0879-2. Epub 2017 Jul 14. PMID: 28710743; PMCID: PMC5511117.
- Cantón R, González-Alba JM, Galán JC. CTX-M Enzymes: Origin and Diffusion. Front Microbiol. 2012 Apr 2;3:110. doi: 10.3389/fmicb.2012.00110. PMID: 22485109; PMCID: PMC3316993.
- Poirel L, Héritier C, Tolün V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004 Jan;48(1):15-22. doi: 10.1128/AAC.48.1.15-22.2004. PMID: 14693513; PMCID: PMC310167.
- Carrër A, Poirel L, Yilmaz M, Akan OA, Feriha C, Cuzon G, Matar G, Honderlick P, Nordmann P. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob Agents Chemother. 2010 Mar;54(3):1369-73. doi: 10.1128/AAC.01312-09. Epub 2010 Jan 19. PMID: 20086157; PMCID: PMC2825965.
- Nordmann P, Naas T, Poirel L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011 Oct;17(10):1791-8. doi: 10.3201/eid1710.110655. PMID: 22000347; PMCID: PMC3310682.
- N G M, C Math G, Nagshetty K, Patil SA, Gaddad SM, Shivannavar CT. Antibiotic Susceptibility Pattern of ESβL Producing Klebsiella pneumoniae Isolated from Urine Samples of Pregnant Women in Karnataka. J Clin Diagn Res. 2014 Oct;8(10):DC08-11. doi: 10.7860/JCDR/2014/9594.5048. Epub 2014 Oct 20. PMID: 25478341; PMCID: PMC4253159.
- Kiratisin P, Apisarnthanarak A, Laesripa C, Saifon P. Molecular characterization and epidemiology of extended-spectrum-beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrob Agents Chemother. 2008 Aug;52(8):2818-24. doi: 10.1128/AAC.00171-08. Epub 2008 May 27. PMID: 18505851; PMCID: PMC2493136.
- Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012 May;18(5):263-72. doi: 10.1016/j.molmed.2012.03.003. Epub 2012 Apr 3. PMID: 22480775.
- Carattoli A. Animal reservoirs for extended spectrum beta-lactamase producers. Clin Microbiol Infect. 2008 Jan;14 Suppl 1:117-23. doi: 10.1111/j.1469-0691.2007.01851.x. Erratum in: Clin Microbiol Infect. 2008 May;14 Suppl 5:21-4. PMID: 18154535.
- Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, Platteel T, Fluit AC, van de Sande-Bruinsma N, Scharinga J, Bonten MJ, Mevius DJ; National ESBL surveillance group. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect. 2011 Jun;17(6):873-80. doi: 10.1111/j.1469-0691.2011.03497.x. Epub 2011 Apr 4. PMID: 21463397.
- Saïdani M, Hammami S, Kammoun A, Slim A, Boutiba-Ben Boubaker I. Emergence of carbapenem-resistant OXA-48 carbapenemase-producing Enterobacteriaceae in Tunisia. J Med Microbiol. 2012 Dec;61(Pt 12):1746-1749. doi: 10.1099/jmm.0.045229-0. Epub 2012 Aug 23. PMID: 22918869.
- Chin TL, McNulty C, Beck C, MacGowan A. Antimicrobial resistance surveillance in urinary tract infections in primary care. J Antimicrob Chemother. 2016 Oct;71(10):2723-8. doi: 10.1093/jac/dkw223. Epub 2016 Jun 26. PMID: 27353470.
- O'Hara CM, Rhoden DL, Miller JM. Reevaluation of the API 20E identification system versus conventional biochemicals for identification of members of the family Enterobacteriaceae: a new look at an old product. J Clin Microbiol. 1992 Jan;30(1):123-5. doi: 10.1128/jcm.30.1.123-125.1992. PMID: 1734043; PMCID: PMC265006.
- CLSI. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 26 th informational supplement. Wayne: CLSI 2016.
- Kumar D, Singh AK, Ali MR, Chander Y. Antimicrobial Susceptibility Profile of Extended Spectrum β-Lactamase (ESBL) Producing Escherichia coli from Various Clinical Samples. Infect Dis (Auckl). 2014 Mar 25;7:1-8. doi: 10.4137/IDRT.S13820. PMID: 24847178; PMCID: PMC4024053.
- Jarlier V, Nicolas MH, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988 Jul-Aug;10(4):867-78. doi: 10.1093/clinids/10.4.867. PMID: 3263690.
- Stapleton PD, Shannon KP, French GL. Carbapenem resistance in Escherichia coli associated with plasmid-determined CMY-4 beta-lactamase production and loss of an outer membrane protein. Antimicrob Agents Chemother. 1999 May;43(5):1206-10. doi: 10.1128/AAC.43.5.1206. PMID: 10223937; PMCID: PMC89134.
- Quiles MG, Menezes LC, Bauab KC, Gumpl EK, Rocchetti TT, Palomo FS, Carlesse F.(2015).Diagnosis of bacteremia in pediatric oncologic patients by in-house real-time PCR. BMC Infect Dis. :15:283.
- Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B, Strömbom L, Ståhlberg A, Zoric N. The real-time polymerase chain reaction. Mol Aspects Med. 2006 Apr-Jun;27(2-3):95-125. doi: 10.1016/j.mam.2005.12.007. Epub 2006 Feb 3. PMID: 16460794.
- Mumbula E, Kwenda G, Samutela M, Kalonda A, Mwansa J, Mwenya D, Koryolova L, Kaile T, Marimo C, Lukwesa –Musyani C. Extended spectrum b-lactamases producing Klebsiella pneumoniae from the neonatal intensive care unit at the university teaching hospital in Lusaka, Zambia. J Med Sci Technol. 2015;4:85–91.
- Caroff N, Espaze E, Bérard I, Richet H, Reynaud A. Mutations in the ampC promoter of Escherichia coli isolates resistant to oxyiminocephalosporins without extended spectrum beta-lactamase production. FEMS Microbiol Lett. 1999 Apr 15;173(2):459-65. doi: 10.1111/j.1574-6968.1999.tb13539.x. PMID: 10227175.
- Cantón R, Novais A, Valverde A, Machado E, Peixe L, Baquero F, Coque TM. Prevalence and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect. 2008 Jan;14 Suppl 1:144-53. doi: 10.1111/j.1469-0691.2007.01850.x. Erratum in: Clin Microbiol Infect. 2008 May;14 Suppl 5:21-4. PMID: 18154538.
- Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011 Jan;66(1):1-14. doi: 10.1093/jac/dkq415. Epub 2010 Nov 16. PMID: 21081548.
- Al-Agamy MH, Shibl AM, Hafez MM, Al-Ahdal MN, Memish ZA, Khubnani H. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli in Riyadh: emergence of CTX-M-15-producing E. coli ST131. Ann Clin Microbiol Antimicrob. 2014 Jan 7;13:4. doi: 10.1186/1476-0711-13-4. PMID: 24397567; PMCID: PMC3898780.
- Rezai MS, Salehifar E, Rafiei A, Langaee T, Rafati M, Shafahi K, Eslami G. Characterization of Multidrug Resistant Extended-Spectrum Beta-Lactamase-Producing Escherichia coli among Uropathogens of Pediatrics in North of Iran. Biomed Res Int. 2015;2015:309478. doi: 10.1155/2015/309478. Epub 2015 May 3. PMID: 26064896; PMCID: PMC4433631.
- ElBouamri MC, Arsalane L, El Kamouni Y, Zouhair S. Antimicrobial susceptibility of urinary Klebsiella pneumoniae and the emergence of carbapenem-resistant strains: A retrospective study from a university hospital in Morocco, North Africa. African Journal of Urology. 2015; 1110-5704.
- Bazzaz BS, Naderinasab M, Mohamadpoor AH, Farshadzadeh Z, Ahmadi S, Yousefi F. The prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae among clinical isolates from a general hospital in Iran. Acta Microbiol Immunol Hung. 2009 Mar;56(1):89-99. doi: 10.1556/AMicr.56.2009.1.7. PMID: 19388560.
- Elhani D, Bakir L, Aouni M, Passet V, Arlet G, Brisse S, Weill FX. Molecular epidemiology of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae strains in a university hospital in Tunis, Tunisia, 1999-2005. Clin Microbiol Infect. 2010 Feb;16(2):157-64. doi: 10.1111/j.1469-0691.2009.03057.x. Epub 2009 Sep 21. PMID: 19769601.
- Mamlouk K, Boutiba-Ben Boubaker I, Gautier V, Vimont S, Picard B, Ben Redjeb S, Arlet G. Emergence and outbreaks of CTX-M beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae strains in a Tunisian hospital. J Clin Microbiol. 2006 Nov;44(11):4049-56. doi: 10.1128/JCM.01076-06. Epub 2006 Sep 6. PMID: 16957046; PMCID: PMC1698301.
- Ferjani S, Saidani M, Ennigrou S, Hsairi M, Slim AF, Ben Boubaker IB. Multidrug resistance and high virulence genotype in uropathogenic Escherichia coli due to diffusion of ST131 clonal group producing CTX-M-15: an emerging problem in a Tunisian hospital. Folia Microbiol (Praha). 2014 May;59(3):257-62. doi: 10.1007/s12223-013-0292-0. Epub 2013 Nov 21. PMID: 24258848.
- Lahlaoui H, Anis BH, Mohamed K, Mohamed BM. Emergence of SHV-12 extended spectrum beta-lactamase among clinical isolates of Enterobacter cloacae in Tunisia. Microb Pathog. 2012 Aug;53(2):64-5. doi: 10.1016/j.micpath.2012.04.003. Epub 2012 Apr 21. PMID: 22543154.
- Agabou A, Pantel A, Ouchenane Z, Lezzar N, Khemissi S, Satta D, Sotto A, Lavigne JP. First description of OXA-48-producing Escherichia coli and the pandemic clone ST131 from patients hospitalised at a military hospital in Algeria. Eur J Clin Microbiol Infect Dis. 2014 Sep;33(9):1641-6. doi: 10.1007/s10096-014-2122-y. Epub 2014 May 5. PMID: 24792128.
- Ben Slama K, Ben Sallem R, Jouini A, Rachid S, Moussa L, Sáenz Y, Estepa V, Somalo S, Boudabous A, Torres C. Diversity of genetic lineages among CTX-M-15 and CTX-M-14 producing Escherichia coli strains in a Tunisian hospital. Curr Microbiol. 2011 Jun;62(6):1794-801. doi: 10.1007/s00284-011-9930-4. Epub 2011 Apr 10. PMID: 21479796.
- Gomi R, Matsuda T, Matsumura Y, Yamamoto M, Tanaka M, Ichiyama S, Yoneda M. Occurrence of Clinically Important Lineages, Including the Sequence Type 131 C1-M27 Subclone, among Extended-Spectrum-β-Lactamase-Producing Escherichia coli in Wastewater. Antimicrob Agents Chemother. 2017 Aug 24;61(9):e00564-17. doi: 10.1128/AAC.00564-17. PMID: 28630184; PMCID: PMC5571296.
- Girlich D, Bouihat N, Poirel L, Benouda A, Nordmann P. High rate of faecal carriage of extended-spectrum β-lactamase and OXA-48 carbapenemase-producing Enterobacteriaceae at a university hospital in Morocco. Clin Microbiol Infect. 2014 Apr;20(4):350-4. doi: 10.1111/1469-0691.12325. Epub 2013 Aug 9. PMID: 23927757.
- Khalaf NG, Eletreby MM, Hanson ND. Characterization of CTX-M ESBLs in Enterobacter cloacae, Escherichia coli and Klebsiella pneumoniae clinical isolates from Cairo, Egypt. BMC Infect Dis. 2009 Jun 4;9:84. doi: 10.1186/1471-2334-9-84. PMID: 19497111; PMCID: PMC2701952.
- Dortet L, Poirel L, Nordmann P. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob Agents Chemother. 2012 Dec;56(12):6437-40. doi: 10.1128/AAC.01395-12. Epub 2012 Oct 15. PMID: 23070158; PMCID: PMC3497194.
- Ktari S, Mnif B, Louati F, Rekik S, Mezghani S, Mahjoubi F, Hammami A. Spread of Klebsiella pneumoniae isolates producing OXA-48 β-lactamase in a Tunisian university hospital. J Antimicrob Chemother. 2011 Jul;66(7):1644-6. doi: 10.1093/jac/dkr181. Epub 2011 May 11. PMID: 21565807.
- Mhaya A, Trabelsi R, M'Zali F, Bégu D, Tounsi S, Gdoura R, Arpin C. First description of NDM-1-positive Klebsiella pneumoniae in the Tunisian community. J Glob Antimicrob Resist. 2020 Jun;21:49-50. doi: 10.1016/j.jgar.2020.02.023. Epub 2020 Mar 10. PMID: 32169682.
- Charfi K, Mansour W, Khalifa AB, Mastouri M, Aouni M, Mammeri H. Emergence of OXA-204 β-lactamase in Tunisia. Diagn Microbiol Infect Dis. 2015 Aug;82(4):314-7. doi: 10.1016/j.diagmicrobio.2015.04.003. Epub 2015 Apr 24. PMID: 26001616.