More Information
Submitted: March 21, 2024 | Approved: April 05, 2024 | Published: April 08, 2024
How to cite this article: Chen J, Wang G, Liang H, Zhao Y, Gao X, et al. Effect of Methyl Jasmonate on the Expression of Transcription Factors in Wild Jujube Seedlings under Salt Stress. Arch Biotechnol Biomed. 2024; 8: 003-008.
DOI: 10.29328/journal.abb.1001038
Copyright License: © 2024 Chen J, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Ziziphus jujuba var. spinosa; Transcription factor; Methyl Jasmonate
Effect of Methyl Jasmonate on the Expression of Transcription Factors in Wild Jujube Seedlings under Salt Stress
Jianing Chen, Guangping Wang, Hanyun Liang, Yan Zhao, Xin Gao, Xiankuan Li* and Jian Zhang*
Traditional Chinese Medicine Resources Laboratory, Tianjin University of Traditional Chinese Medicine, Tianjin 301617, China
*Address for Correspondence: Jian Zhang, Traditional Chinese Medicine Resources Laboratory, Tianjin University of Traditional Chinese Medicine, Tianjin 301617, China, Email: [email protected]
Xiankuan Li, Traditional Chinese Medicine Resources Laboratory, Tianjin University of Traditional Chinese Medicine, Tianjin 301617, China, Email: [email protected]
Methyl Jasmonate (MeJA) can be used as a signal molecule to regulate the expression of resistance genes in the resistance to abiotic stress, thus improving the salt tolerance of wild jujube. Among the resistance genes combined with methyl jasmonate, transcription factors play an important role in response to salt stress. However, the interaction of transcription factors in different tissues under salt stress and the regulation of transcription factors by MeJA remain unclear. In this study, the effects of MeJA on transcription factor expression in wild jujube under salt stress were investigated, and the differences in transcription factor expression among different tissues were compared. It was found that MeJA could increase the type and quantity of transcription factors responding to salt stress. The types of transcription factors responding to salt stress were roughly the same among different tissues, but the quantity and expression of the transcription factors were significantly different. The results of transcription factor co-expression analysis showed that transcription factors play synergistic roles in the face of abiotic stress, which can provide preferable genes for subsequent transgenic work.
Among abiotic stresses, high salt stress is the most serious environmental stress [1], affecting at least 20% of irrigated land worldwide. When salt ions in plants accumulate excessively, the normal growth and development of plants will be affected. To cope with the damage caused by these salt stresses, plants have also developed some coping strategies. Under high salt stress, the expression of a variety of genes is up-regulated, the products of which are directly or indirectly involved in plant protection, such as some genes encoding osmofactors, ion channels, receptors, calcium signaling components, and some regulatory signaling factors or enzymes, which can be transferred to sensitive plants to develop a salt-tolerant phenotype.
Transcription factors play an important role in the plant's response to salt stress. For example, the bZIP transcription factor TabZIP 15 can improve the tolerance of wheat to salt stress [2]; the AvNAC 030 plays a positive role in the salt tolerance regulation mechanism in kiwifruit [3]; the Transcriptome analysis of eggplant under salt stress showed that the AP2/ERF transcription factor SmERF 1 was a positive regulator of salt stress [4]; and the bHLH transcription factor AhbHLH 121 can improve the salt tolerance of peanuts [5]. In addition to the above-mentioned transcription factors that can positively regulate plant salt tolerance, some can also play a negative role. For example, the transcription factor GmERF105 negatively regulates salt tolerance in Arabidopsis [6].
There is a close relationship between Jasmonic Acid (JA) and salt stress [7-12]. As an endogenous signaling molecule, JA can participate in the abiotic response of plants to salt stress. Jasmonic acid and its methylated derivatives methyl jasmonate and amino acid derivatives are collectively referred to as jasmonic acid compounds. (JAs) [13]. JAs are not only related to their growth and development but also to the plant defense system. It has been found that JAs have a wide range of physiological effects on plant growth. It can not only affect seed germination, plant growth and development, photosynthesis, and other processes, but also play a role as a signal molecule in the process of resistance to biological and abiotic stress, regulating the expression of genes related to stress products in cells, and promoting the accumulation of secondary metabolites [14].
Wild jujube (Ziziphus jujuba var. spinosa), a woody plant belonging to the rhamnus family, can be used as medicine [15-17]. Wild jujube likes warm and dry climates; the plant itself has a strong tolerance to cold, drought, salt, and alkali. When the wild jujube is subjected to low salt stress, its good tolerance will not only not hurt the plant, but also increase the accumulation of its medicinal ingredients.
This study highlights the effect of MeJA on the expression of transcription factors in wild jujube under salt stress, to explore how wild jujube responds to salt stress, the level of transcription factors after salt stress, and what changes the transcription factors will encounter when single salt stress is alleviated by MeJA. In addition, this study also compared the different transcription factors in the roots and leaves of wild jujube seedlings under the same stress conditions, providing ideal experimental materials for subsequent experiments.
Experimental materials
a suitable amount of full and uniform wild jujube seeds were selected and evenly placed in petri dishes lined with moist filter paper, and germination was carried out at 25 °C, during which a moist environment was maintained all the time. When the seeds sprouted, they were evenly planted into 50-hole seedling hole trays with a seedling substrate of vermiculite: perlite (1:1). Then they were placed in the incubation room with a 16L:8D photoperiod, a light intensity of 3600lx, a constant temperature of 23 (soil 2) ℃, relative humidity of (50+5)% and watered regularly. After forty days, the seedlings were divided into two groups. Group one (N) and group two (M) were stressed with 150 mmol/L salt concentration for ten days. After ten days, group one was cultured normally, and group two continued to be foliar sprayed with 200 μmmol/L MeJA.
Transcriptome sequencing
Total RNA extraction, cDNA library construction, and sequencing of all samples in this experiment were done by PersonalBio, Shanghai. Total RNA extraction for each sample was performed using Trizol reagent, which is suitable for the rapid isolation of RNA from tissues and cells and maintains the integrity of RNA during cell breakage and lysis. After obtaining total RNA of qualified quality, mRNA with a polyA structure in total RNA was enriched by Oligo(dT) magnetic beads, and ionic interruption was used to interrupt the RNA to a fragment of about 300 bp in length. The first strand of cDNA was synthesized using RNA as a template with a 6-base random primer and reverse transcriptase, and the second strand of cDNA was synthesized using the first strand of cDNA as a template. After the library construction was completed, PCR amplification was used for library fragment enrichment, followed by library selection based on the fragment size, which was 450 bp. Next, the library was subjected to quality control by an Agilent 2100 Bioanalyzer, and then the total concentration of the library and the effective concentration of the library were detected. Then, according to the effective concentration of the library and the amount of data required for the library, the libraries containing different index sequences (each sample plus a different index, and finally separating the downstream data of each sample according to the index) were mixed proportionally. The mixed libraries were uniformly diluted to 2 nM and denatured by base denaturation to form single-stranded libraries. After the samples were RNA extracted, purified, and constructed into libraries, these libraries were subjected to double-end (Paired-end, PE) sequencing using Next-GenerationSequencing (NGS) technology based on the Illumina sequencing platform.
No reference transcriptome analysis process
The original disembarking data were FASTQ files. First, the original disembarking data was filtered, and Reads with connectors, less than 50 bp in length and average sequence quality below Q20 were removed. The resulting high-quality sequences were splintered from scratch to obtain transcript sequences, and the transcripts were clustered. The longest transcript was selected as Unigene, and then Unigene was used for subsequent GO, KEGG, eggNOG, SwissProt, Pfam annotation, ORF prediction, SSR prediction, etc. At the same time, the filtered sequences were compared to the Unigene to obtain the Reads Count of each Unigene. On this basis, the sample was further analyzed for expression difference analysis, enrichment analysis, cSNP analysis, and InDel analysis.
Analysis of gene expression level
Using the transcriptome expression quantification software RSEM, Clean Reads of each sample were compared to the reference sequence using the transcript sequence as a reference. Then the number of Reads from each sample to each gene was counted, and the FPKM value of each gene was calculated.
Transcription factor analysis
transcription factor prediction is done by comparing plants and animals with PlantTFDB (Plant Transcription Factor Database) and AnimalTFDB (Animal Transcription Factor Data Base) databases respectively, to predict the transcription factor and the family information to which the transcription factor belongs.
Construction of coexpression network of differential transcription factors in root and leaf of wild jujube: Excel was used to sort out the expression data of differential genes. SPSS was used to analyze the correlation between the expression of differential transcription factors in wild jujube leaf and root. Use Chiplot for data visualization. Analysis of visual results: The darker the color and the larger the square, the stronger the correlation.
Data processing
Calculations and organization of test results using Excel 2019; data analysis using SPSS 21.0; and graphing using Excel and Chiplot.
Comparative analysis of transcription factors
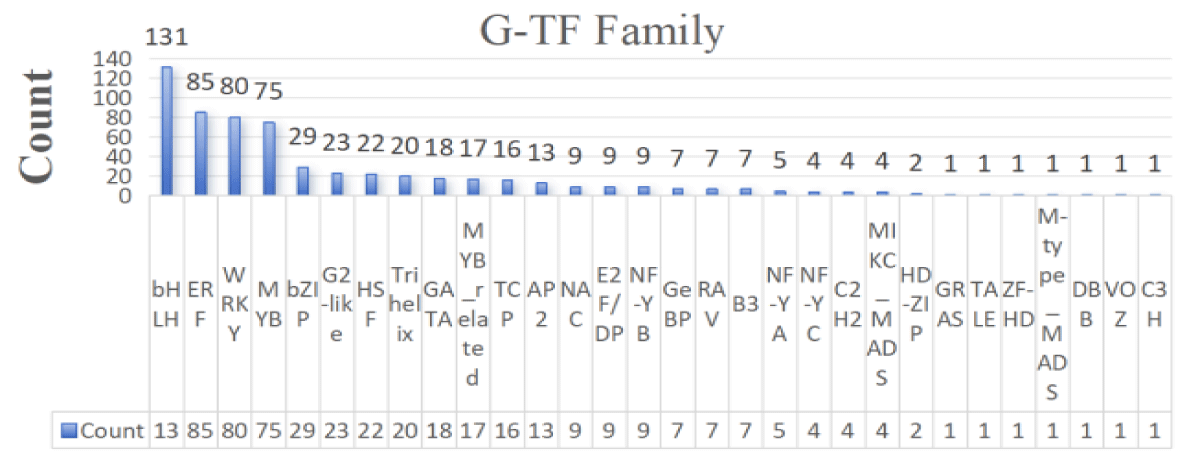
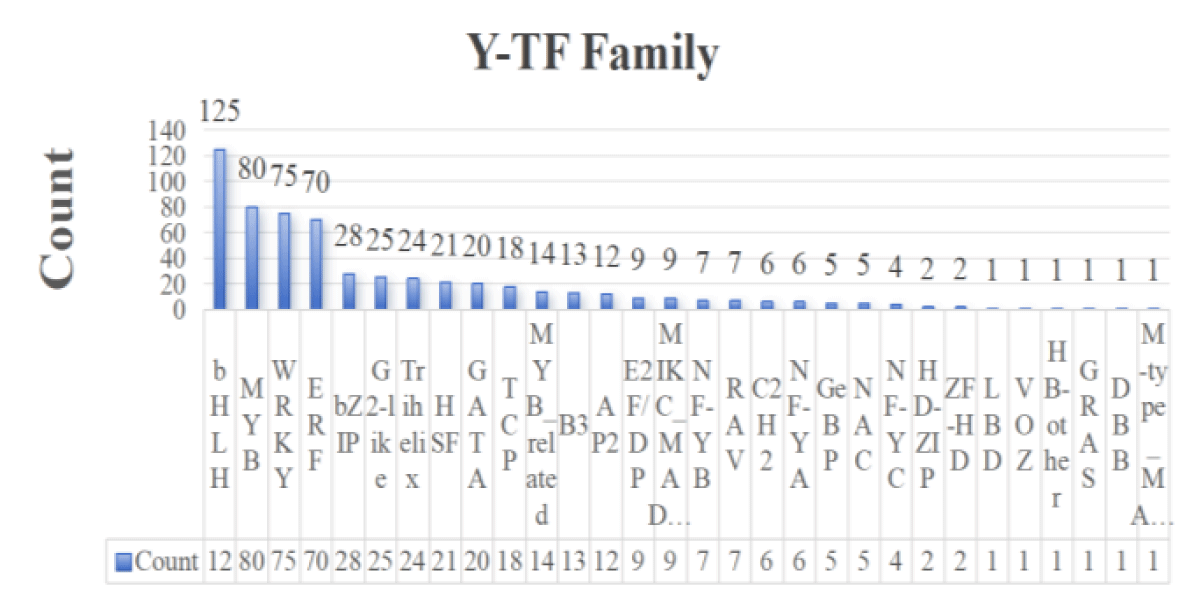
A total of 1,196 transcription factors from 30 gene families, including bHLH, NAC, ERF, C2H2, bZIP, WRKY, and MYB, were identified from the transcriptome sequencing data. Among them, 603 transcription factors were found in roots and 593 in leaves (Figures 1,2). Combined with the type and distribution of transcription factors, the bHLH family had the most transcription factors in roots, followed by the ERF family, and seven families, including GRAS, had only one transcription factor; in leaves, the bHLH family also had the most transcription factors, but the MYB family had the second-highest number of transcription factors, and the same six families, GRAS and TALE, had only one transcription factor.
Figure 1: Statistical map of root-transcription factor families.
Figure 2: Statistical map of leaf-transcription factor families.
The above transcription factors were screened on the principle of |log2FoldChange|>1 and significance p – value < 0.05, and the screened differential genes were subjected to subsequent analyses. The differentially expressed transcription factors in the roots and leaves of wild jujube under salt stress and MeJA alleviation are shown in Figures 3,4. In terms of the number of transcription factors, the number of differential genes was greater in roots than in leaves, suggesting that plant roots were preferentially exposed to the stress environment and responded to a greater number of transcription factors in response to longer exposure to salt stress than did leaves. In terms of the types of transcription factors, the highest number of transcription factors of the ERF family was found in roots, whereas the highest number of transcription factors of the WRKY family was found in leaves, suggesting that transcription factors play different roles in different tissues in the face of salt stress.
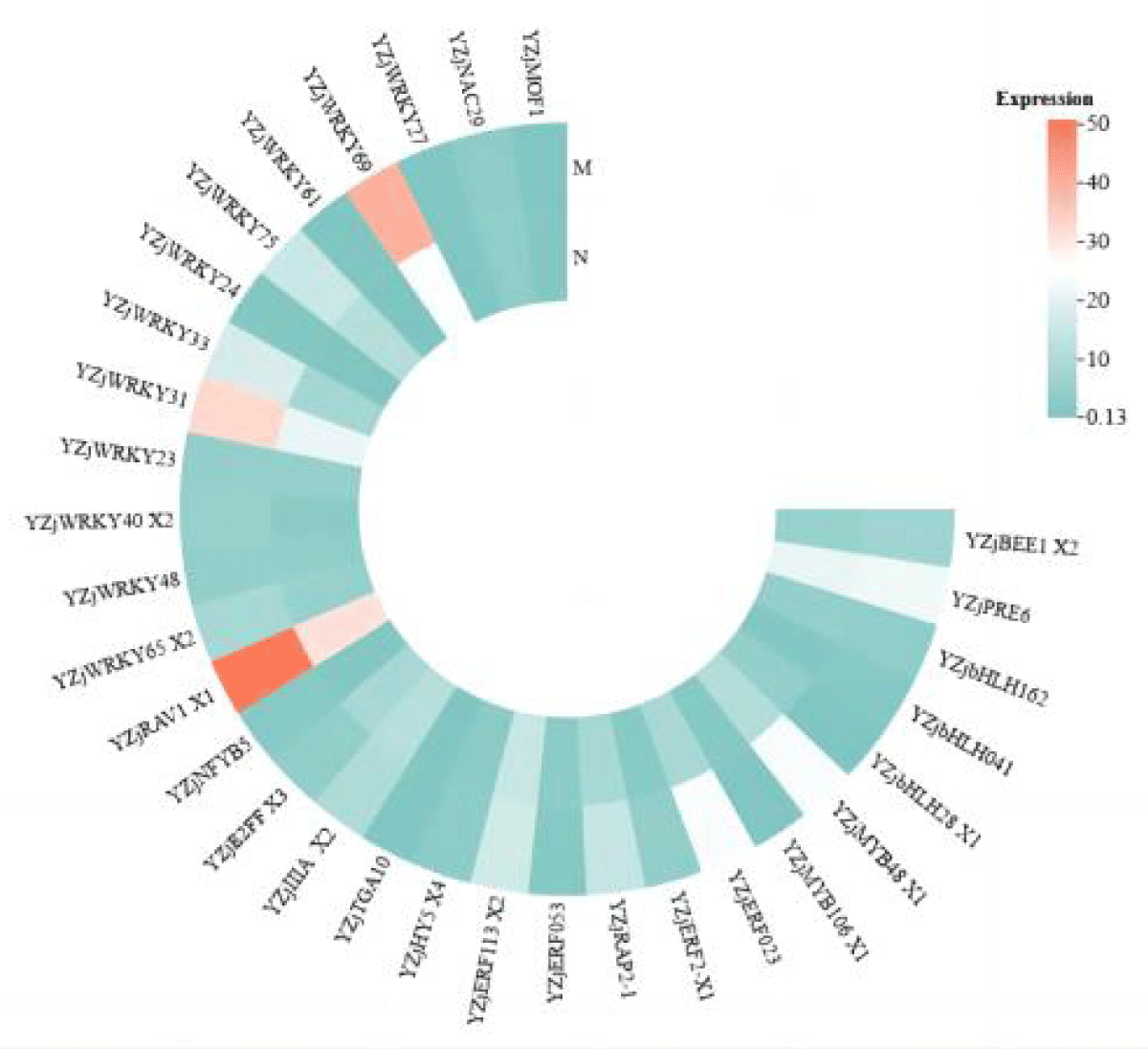
As can be seen in Figure 3, the expression of most transcription factor family genes was significantly up-regulated after leaf salt stress was alleviated by MeJA, and the expression of the genes alleviated by MeJA was higher than that of the normal salt stress group. In the leaves, the number of differentially expressed transcription factors of three families, WRKY, bHLH, and ERF, was high. Among them, the expression of a total of 12 transcription factors in the WRKY family was up-regulated. ZjWRKY 31 and ZjWRKY 69 were not only significantly different but also highly expressed. 5 genes in the ERF family, including ZjERF 023 and ZjERF 053, were all up-regulated.
Figure 3: Heat map of expression of various transcription factors in leaves after alleviation by MeJA.
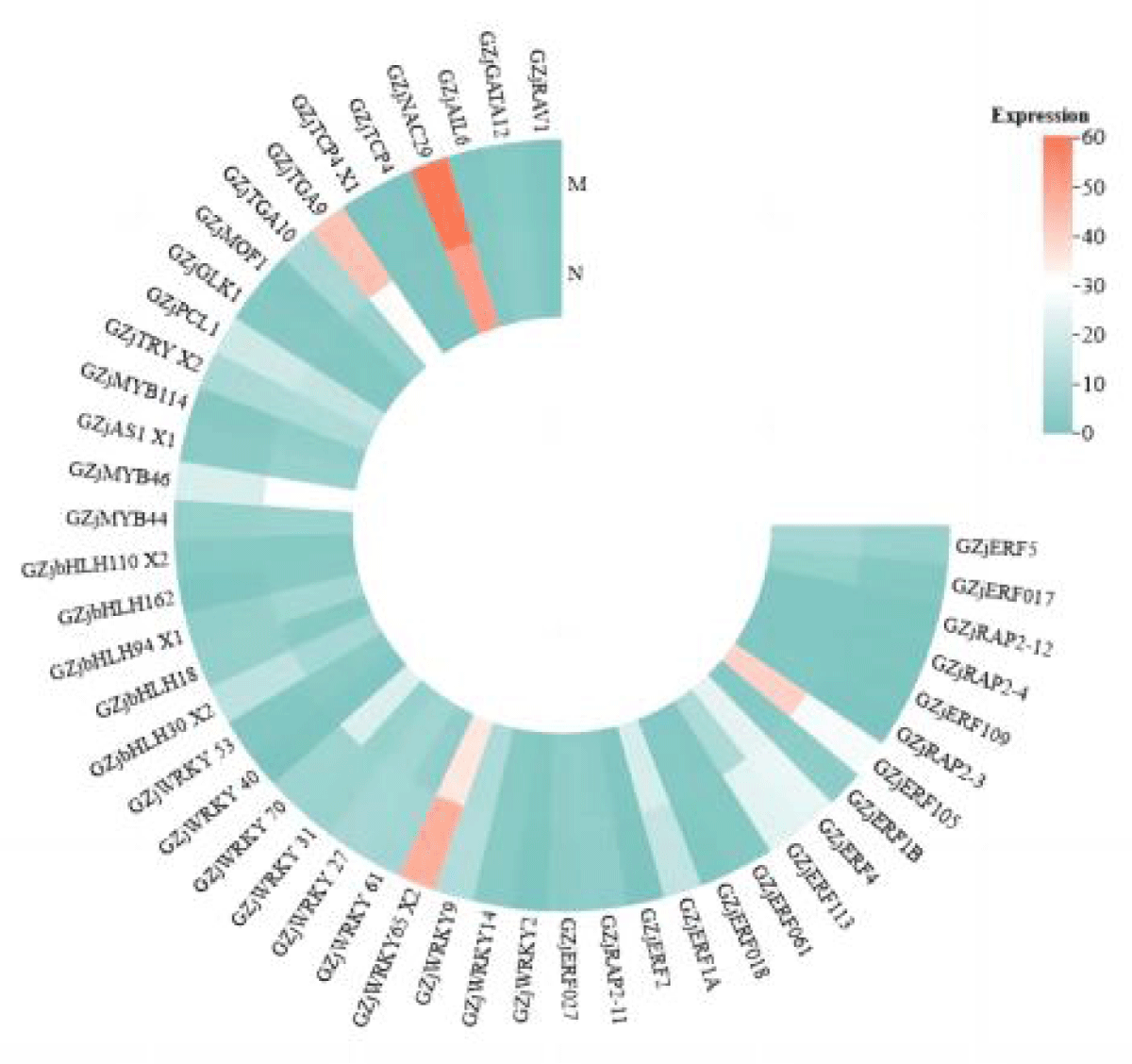
As can be seen in Figure 4, unlike the expression of transcription factors in leaves, the expression of most of the transcription factor family members was predominantly down-regulated in roots, and the difference between the single salt stress group and the MeJA-relieved group was not significant. As in leaves, the number of differentially expressed transcription factors of the WRKY, bHLH, and ERF families was high, unlike in roots, where the number of ERF family members was higher after relief by MeJA. The expression of all WRKY and ERF family transcription factors was up-regulated in leaves, whereas a total of six transcription factors of the WRKY family, ZjWRKY 14, ZjWRKY 29, ZjWRKY 27 et., were up-regulated for expression in roots, and ZjWRKY 2, ZjWRKY 53 et., a total of five genes were down-regulated, and the ratio of the number of up-regulated and down-regulated gene expression was almost equal; all ERF family members were down-regulated except for three genes, ZjERF 1B, ZjERF 113, and ZjRAP 2-11, which was extremely different from the situation in leaves. It can be seen that the expression of transcription factors varies greatly depending on the plant tissue. In addition, the expression heatmap shows that transcription factors in roots are not only more diverse and numerous than those in leaves but also have higher expression than that of transcription factors in leaves.
Figure 4: Heat map of expression of various transcription factors in roots after alleviation by MeJA.
According to Figures 3,4, it can be seen that ZjWRKY 31 and ZjWRKY 27 of the WRKY family, ZjTGA 10 of the bZIP family, ZjNAC 29 of the NAC family, and ZjMOF 1 of the G2-like family play roles in both roots and leaves and their expression is shown to be up-regulated. However, ZjbHLH 162 of the bHLH family, although it plays a regulatory role in both tissues, is up-regulated in leaves but down-regulated in roots.
Transcription factor co-expression analysis
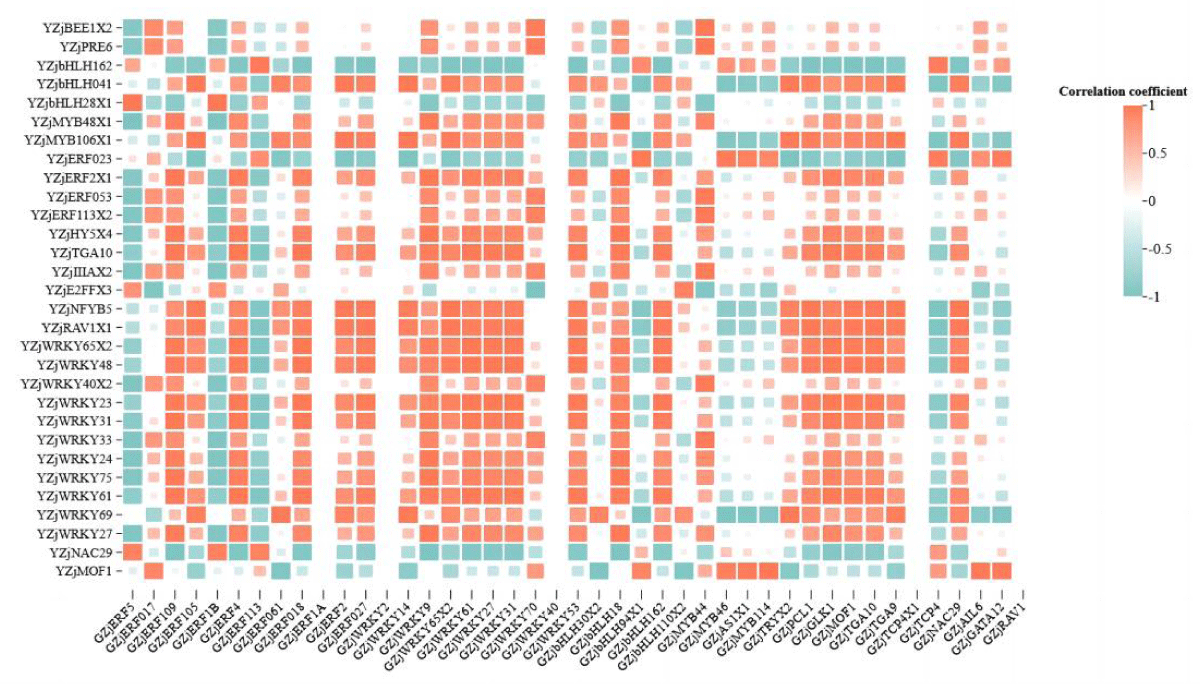
In the differential transcription factor co-expression network in roots and leaves of wild jujube seedlings after salt stress alleviated by MeJA, the correlation between genes was calculated. It was found that most of the genes were positively correlated (Figure 5). For example, GZjWRKY 53 was significantly positively correlated with YZjWRKY 48 (p < 0.01). And a small portion of the genes were negatively correlated with each other, for example, GZjMYB 46 was significantly negatively correlated with YZjWRKY 69, which was significantly negatively correlated (p < 0.01). Some individual genes showed different correlations with different genes; for example, GZjERF 109 was significantly positively correlated with YZjWRKY 31 (p < 0.01), and significantly negatively correlated with YZjNAC 29 (p < 0.05). Some genes were significantly correlated with several genes at the same time, and all of them were positively correlated. For example, GZjERF 018, GZjWRKY 61, and GZjGLK1 were significantly positively correlated with YZjWRKY 65 isoformX2, YZjWRKY 23, and YZjWRKY 48 (p < 0.05).
Figure 5: Heat map of correlation of differential transcription factors in roots and leaves. Note: Horizontal is root differential transcription factor, vertical is leaf differential transcription factor.
Wild jujube (Ziziphus jujuba var. spinosa) is a woody plant of the genus Ziziphus in the family Rhamnaceae. Native to China, wild jujube has a certain degree of salt tolerance. Other studies have shown that MeJA can induce disease-resistant and insect-resistant responses in plants and improve their stress tolerance [18,19]. In this study, we used the salt-tolerant wild jujube seedlings as experimental materials, which were successively induced by salt stress and relieved by MeJA, to investigate the different responses of different tissues of wild jujube to MeJA at the level of transcription factors.
Comparative analysis of transcription factors in roots and leaves revealed that the family of transcription factors that played the main coordinating role was approximately the same in different tissues after salt stress, but the families that worked in concert with each other differed depending on the tissue. Analysis of differential transcription factors showed that the number of differential genes was greater in roots than in leaves, assuming that plant roots were preferentially exposed to the stress environment and for a longer time than leaves were exposed to salt stress, resulting in a greater number of responsive transcription factors. Similarly, the analysis showed that the expression of transcription factors was higher in the MeJA-relieved group than in the single salt stress group in leaves, whereas the difference between the two groups was not significant in roots, suggesting that transcription factors in leaves were more sensitive to MeJA due to the preferential and longer exposure of leaves to MeJA as a result of applying MeJA by foliar spraying. The increase in the expression of transcription factors after leaf surface spraying with MeJA also indicates that MeJA may regulate physiological processes through transcription factors at the molecular level to alleviate the harm caused by salt stress on plants. Furthermore, it provides the basis for methyl jasmonate to improve salt stress.
The trend of gene changes showed that ERF was fully up-regulated in leaves but almost fully down-regulated in roots. This may be related to the physiological regulation of plants by ethylene: when the plant is subjected to abiotic stress, ethylene becomes an important substance in inducing senescence and abscission of leaves [20,21].
Specific analyses of differential transcription factors in roots and leaves revealed that 1. the types of transcription factors that play major coordinating roles are roughly the same in roots and leaves, but the members of each family that play roles in different tissues are different; 2. there are a few transcription factors that play roles at the same time in roots and leaves, with the same trend of change in expression. What our research has found is that GZjWRKY 53 was significantly positively correlated with YZjWRKY 48 (p < 0.01). This suggests that GZjWRKY 53 and YZjWRKY 48 play a synergistic role in the face of stress [22-25]. 3. the transcription factors that play roles at the same time in roots and leaves have different trends of change in expression. For example, ZjbHLH 162 exhibits functional significance in both root and leaf tissues, albeit with distinct expression patterns, implying that diverse transcription factors may employ tissue-specific mechanisms of action. Co-expression analyses of the differential genes more strongly confirmed that the transcription factors act synergistically in the face of abiotic stresses. Studying the co-expression of transcription factors can provide preferable genes for subsequent transgenic work.
- Tuteja N. Mechanisms of high salinity tolerance in plants. Methods Enzymol. 2007;428:419-38. doi: 10.1016/S0076-6879(07)28024-3. PMID: 17875432.
- Bi C, Yu Y, Dong C, Yang Y, Zhai Y, Du F, Xia C, Ni Z, Kong X, Zhang L. The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat. Plant Biotechnol J. 2021 Feb;19(2):209-211. doi: 10.1111/pbi.13453. Epub 2020 Aug 13. PMID: 32702168; PMCID: PMC7868967.
- Li M, Wu Z, Gu H, Cheng D, Guo X, Li L, Shi C, Xu G, Gu S, Abid M, Zhong Y, Qi X, Chen J. AvNAC030, a NAC Domain Transcription Factor, Enhances Salt Stress Tolerance in Kiwifruit. Int J Mol Sci. 2021 Nov 2;22(21):11897. doi: 10.3390/ijms222111897. PMID: 34769325; PMCID: PMC8585034.
- Shen L, Zhao E, Liu R, Yang X. Transcriptome Analysis of Eggplant under Salt Stress: AP2/ERF Transcription Factor SmERF1 Acts as a Positive Regulator of Salt Stress. Plants (Basel). 2022 Aug 25;11(17):2205. doi: 10.3390/plants11172205. PMID: 36079586; PMCID: PMC9460861.
- Zhao X, Wang Q, Yan C, Sun Q, Wang J, Li C, Yuan C, Mou Y, Shan S. The bHLH transcription factor AhbHLH121 improves salt tolerance in peanut. Int J Biol Macromol. 2024 Jan;256(Pt 2):128492. doi: 10.1016/j.ijbiomac.2023.128492. Epub 2023 Nov 28. PMID: 38035960.
- Li L, Zhu Z, Liu J, Zhang Y, Lu Y, Zhao J, Xing H, Guo N. Transcription Factor GmERF105 Negatively Regulates Salt Stress Tolerance in Arabidopsis thaliana. Plants (Basel). 2023 Aug 21;12(16):3007. doi: 10.3390/plants12163007. PMID: 37631217; PMCID: PMC10459988.
- Aliakbari M, Tahmasebi S, Sisakht JN. Jasmonic acid improves barley photosynthetic efficiency through a possible regulatory module, MYC2-RcaA, under combined drought and salinity stress. Photosynth Res. 2024 Jan;159(1):69-78. doi: 10.1007/s11120-023-01074-2. Epub 2024 Feb 8. PMID: 38329704.
- Dhaka P, Tallapragada S, Devi S. Implication of Jasmonic Acid on Physiological Alterations on Salt Stressed Fodder Sorghum (Sorghum bicolor L.). International Journal of Environment and Climate Change,2023,13(8):649-660.
- Simon N, Hieu TN, Elisabeth E. Jasmonate signaling controls negative and positive effectors of salt stress tolerance in rice.Journal of experimental botany. 2023; 74(10):
- Gomez MY, Schroeder MM, Chieb M, McLain NK, Gachomo EW. Bradyrhizobium japonicum IRAT FA3 promotes salt tolerance through jasmonic acid priming in Arabidopsis thaliana. BMC Plant Biology. 2023; 23(1):60-60.
- Sheteiwy MS, Ulhassan Z, Qi W, Lu H. Association of jasmonic acid priming with multiple defense mechanisms in wheat plants under high salt stress#13. Frontiers in Plant Science. 2022;13886862-886862.
- Feng RS, Ting TL, Cheng WL. Jasmonic Acid Impairs Arabidopsis Seedling Salt Stress Tolerance Through MYC2-Mediated Repression of CAT2 Expression#13. Frontiers in Plant Science,2021,12730228-730228.
- Jinjing P, Houping W, Qiugui Y. Jasmonate-regulated seed germination and crosstalk with other phytohormones. Journal of experimental botany. 2023; 74(4):1162-1175.
- Ma L, Liu X, Lv W, Yang Y. Molecular Mechanisms of Plant Responses to Salt Stress. Front Plant Sci. 2022 Jun 27;13:934877. doi: 10.3389/fpls.2022.934877. PMID: 35832230; PMCID: PMC9271918.
- Dong XN, Li MT, Gu HY, Zhu Y, Gu X. Advances in pharmacological effects of jujuboside B. Zhongguo Zhong yao za zhi = Zhongguo zhongyao zazhi = China Journal of Chinese materia medica. 2023;48(16):4295-4301.
- Mei Z, Jinrui L, Yanqing Z. Zizyphi Spinosae Semen: a natural herb resource for treating neurological disorders.[J]. Current topics in medicinal chemistry. 2022; 22(17).
- Ali Z, Saeed C, Behnam M. The physiological and pharmacological effects of Ziziphoratenuior L.: A review study. Avicenna journal of phytomedicine. 2022;12(1):16-29.
- Bhavanam S, Stout M. Seed Treatment With Jasmonic Acid and Methyl Jasmonate Induces Resistance to Insects but Reduces Plant Growth and Yield in Rice, Oryza sativa. Front Plant Sci. 2021 Aug 16;12:691768. doi: 10.3389/fpls.2021.691768. PMID: 34484259; PMCID: PMC8415220.
- Kraus EC, Stout MJ. Seed treatment using methyl jasmonate induces resistance to rice water weevil but reduces plant growth in rice. PLoS One. 2019 Sep 23;14(9):e0222800. doi: 10.1371/journal.pone.0222800. PMID: 31545832; PMCID: PMC6756538.
- Chen W, Huang B. Cytokinin or ethylene regulation of heat-induced leaf senescence involving transcriptional modulation of WRKY in perennial ryegrass. Physiol Plant. 2022 Sep;174(5):e13766. doi: 10.1111/ppl.13766. PMID: 36053893.
- Song S, Ge M, Wang W, Gu C, Chen K, Zhang Q, Yu Q, Liu G, Jiang J. BpEIN3.1 represses leaf senescence by inhibiting synthesis of ethylene and abscisic acid in Betula platyphylla. Plant Sci. 2022 Aug; 321:111330. doi: 10.1016/j.plantsci.2022.111330. Epub 2022 May 25. PMID: 35696929.
- Han, Deguo Xu. Overexpression of MxWRKY53 increased iron and high salinity stress tolerance in Arabidopsis thaliana. In Vitro Cellular Developmental Biology – Plant. 2021; 58(2):1-13.
- Fanwei M, Xunmei Z, Jia W. The GRAS protein OsDLA involves in brassinosteroid signalling and positively regulates blast resistance by forming a module with GSK2 and OsWRKY53 in rice. Plant biotechnology journal. 2023; 22(2):363-378.
- Balfagón D, Pascual SL, Sengupta S. WRKY48 negatively regulates plant acclimation to a combination of high light and heat stress. The Plant Journal: for Cell and Molecular Biology. 2024.
- Xiaolu W, Lulu C, Xinyi L. Integrating physiological and transcriptome analyses clarified the molecular regulation mechanism of PyWRKY48 in poplar under cadmium stress. International Journal of Biological Macromolecules. 2023; 238124072-124072.